Abstract
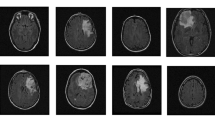
Cervical cancer has influenced life of women worldwide as the fourth most severe cancer. Early screening, detection and treatment of cervical cancer notably increase the life quality and reduce the death rate of patients. Therefore, automatic diagnosis of cervical cancer could bridge the gap between testing needs and capabilities. Cervical cell detection plays an important role in cancer screening, the intent of this study is to classify the cervical cells through deep learning models, which helps to monitor the patients’ health. SSD (Single Shot MultiBox Detector) network is integrated with the positive and negative features to address the shortcomings of insufficient sensitivity to small objects. Besides, center loss function is added to better address situations that intra-class differences are greater than inter-class differences. A dataset containing 1462 benchmarked cervical cells was utilized. 80% (1167) are used for training and the remaining 20% (295) are allocated for testing. Proposed optimized SSD network achieved the accuracy of 90.8% and mAP (mean Average Precision) of 81.53%, which is 7.54% and 4.92% higher than YOLO (You Only Look Once) and classical SSD, respectively. The addition of complementary features improves the network sensitivity and the overall accuracy. It is also concluded that the proposed SSD network could be applied in cell classification for the early automatic detection of cervical cancer.











Similar content being viewed by others
References
Agrawal S, Raidu B, Agrawal K, Barik RC (2017) Prediction of cancerous cell by cluster based biomedical CT image and analysis[J]. IOSR J Dental Med Sci 16(2):67–73
Allehaibi KHS, Nugroho LE, Lazuardi L et al (2019) Segmentation and classification of cervical cells using deep learning[J]. IEEE Access 7:116925–116941
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, Bray F (2020) Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis[J]. Lancet Glob Health 8(2):e191–e203
Arya M, Mittal N, Singh G (2018) Texture-based feature extraction of smear images for the detection of cervical cancer[J]. IET Comput Vis 12(8):1049–1059
Basheer S, Mariyam Aysha Bivi S, Jayakumar S et al (2019) Machine learning based classification of cervical cancer using K-nearest neighbour, random forest and multilayer perceptron algorithms[J]. J Comput Theor Nanosci 16(5–6):2523–2527
Buggenthin F, Marr C, Schwarzfischer M et al (2013) An automatic method for robust and fast cell detection in bright field images from high-throughput microscopy[J]. BMC Bioinformatics 14(1):1–12
Dewi EM, Purwanti E, Apsari R (2019) Cervical Cell Classification using Learning Vector Quantization (LVQ) Based on Shape and Statistical Features[J]. Int J Online Biomed Eng 15(2)
Dong N, Zhao L, Wu CH, Chang JF (2020) Inception v3 based cervical cell classification combined with artificially extracted features[J]. Appl Soft Comput 93:106311
Girshick R, Donahue J, Darrell T et al (2014) Rich feature hierarchies for accurate object detection and semantic segmentation[C]. Proceedings of the IEEE conference on computer vision and pattern recognition, pp 580–587
Gorantla R, Singh R K, Pandey R et al (2019) Cervical cancer diagnosis using cervixnet-a deep learning approach[C]. 2019 IEEE 19th international conference on bioinformatics and bioengineering (BIBE). IEEE, pp 397–404
He K, Gkioxari G, Dollár P et al (2017) Mask r-cnn[C]. Proceedings of the IEEE international conference on computer vision pp 2961–2969
Hu W, Huang Y, Wei L, Zhang F, Li H (2015) Deep convolutional neural networks for hyperspectral image classification[J]. J Sensors 2015:1–12
Huang P, Zhang S, Li M, Wang J, Ma C, Wang B, Lv X (2020) Classification of cervical biopsy images based on lasso and EL-SVM[J]. IEEE Access 8:24219–24228
Iliyasu AM, Fatichah C (2017) A quantum hybrid PSO combined with fuzzy k-NN approach to feature selection and cell classification in cervical cancer detection[J]. Sensors 17(12):2935
Iqbal T, Ali H (2018) Generative adversarial network for medical images (MI-GAN)[J]. J Med Syst 42(11):1–11
Jia AD, Li BZ, Zhang CC (2020) Detection of cervical cancer cells based on strong feature CNN-SVM network[J]. Neurocomputing 411:112–127
Khamparia A, Gupta D, de Albuquerque VHC et al (2020) Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning[J]. J Supercomput:1–19
Kido S, Hirano Y, Hashimoto N (2018) Detection and classification of lung abnormalities by use of convolutional neural network (CNN) and regions with CNN features (R-CNN)[C]. 2018 international workshop on advanced image technology (IWAIT). IEEE, pp 1–4
Kuko M, Pourhomayoun M (2020) Single and clustered cervical cell classification with ensemble and deep learning methods[J]. Inf Syst Front 22(5):1039–1051
Li K, Sun H, Lu Z, Xin J, Zhang L, Guo Y, Guo Q (2018) Value of [18F] FDG PET radiomic features and VEGF expression in predicting pelvic lymphatic metastasis and their potential relationship in early-stage cervical squamous cell carcinoma[J]. Eur J Radiol 106:160–166
Lin H, Hu Y, Chen S, Yao J, Zhang L (2019) Fine-grained classification of cervical cells using morphological and appearance based convolutional neural networks[J]. IEEE Access 7:71541–71549
Lin YC, Lin CH, Lu HY, Chiang HJ, Wang HK, Huang YT, Ng SH, Hong JH, Yen TC, Lai CH, Lin G (2020) Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer[J]. Eur Radiol 30(3):1297–1305
Lu J, Song E, Ghoneim A, Alrashoud M (2020) Machine learning for assisting cervical cancer diagnosis: an ensemble approach[J]. Futur Gener Comput Syst 106:199–205
Malli PK, Nandyal S (2017) Machine learning technique for detection of cervical cancer using k-NN and artificial neural network[J]. Int J Emerg Trends Technol Comput Sci (IJETTCS) 6(4)
Momenimovahed Z, Salehiniya H (2017) Incidence, mortality and risk factors of cervical cancer in the world[J]. Biomed Res Ther 4(12):1795–1811
Rayavarapu K, Krishna KKV (2018) Prediction of cervical cancer using voting and DNN classifiers[C]. 2018 international conference on current trends towards converging technologies (ICCTCT). IEEE, pp 1–5
Redmon J, Farhadi A (2018) Yolov3: An incremental improvement[J]. arXiv preprint arXiv:1804.02767
Ren S, He K, Girshick R et al (2015) Faster r-cnn: Towards real-time object detection with region proposal networks[J]. arXiv preprint arXiv:1506.01497
Saroja B, Priyadharson ASM (2018) Colon Cancer detection methods–a review[J]. Annu Res Rev Biol 24:1–16
Shiraz A, Crawford R, Egawa N, Griffin H, Doorbar J (2020) The early detection of cervical cancer. The current and changing landscape of cervical disease detection[J]. Cytopathology 31(4):258–270
Singh SK, Goyal A (2020) Performance analysis of machine learning algorithms for cervical cancer detection[J]. Int J Healthc Inf Syst Inform (IJHISI) 15(2):1–21
Singh N K, Raza K (2020) Medical Image Generation using Generative Adversarial Networks[J]. arXiv preprint arXiv:2005.10687
Sompawong N, Mopan J, Pooprasert P et al (2019) Automated pap smear cervical cancer screening using deep learning[C]. 2019 41st annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 7044–7048
Sophea P, Handayani D O D, Boursier P (2018) Abnormal cervical cell detection using hog descriptor and SVM classifier[C]. 2018 fourth international conference on advances in computing, Communication & Automation (ICACCA). IEEE, pp 1–6
Sun G, Li S, Cao Y et al (2017) Cervical cancer diagnosis based on random forest[J]. Int J Perform Eng:13(4)
Thohir M, Foeady A Z, Novitasari D C R et al (2020) Classification of colposcopy data using GLCM-SVM on cervical Cancer[C]. 2020 international conference on artificial intelligence in information and communication (ICAIIC). IEEE, pp 373–378
Wentzensen N, Lahrmann B, Clarke MA, Kinney W, Tokugawa D, Poitras N, Locke A, Bartels L, Krauthoff A, Walker J, Zuna R, Grewal KK, Goldhoff PE, Kingery JD, Castle PE, Schiffman M, Lorey TS, Grabe N (2021) Accuracy and efficiency of deep-learning–based automation of dual stain cytology in cervical Cancer screening[J]. JNCI: J Natl Cancer Inst 113(1):72–79
William W, Ware A, Basaza-Ejiri AH, Obungoloch J (2018) A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images[J]. Comput Methods Prog Biomed 164:15–22
Xu T, Zhang H, Xin C, Kim E, Long LR, Xue Z, Antani S, Huang X (2017) Multi-feature based benchmark for cervical dysplasia classification evaluation[J]. Pattern Recogn 63:468–475
Yoo TK, Choi JY, Kim HK (2020) A generative adversarial network approach to predicting postoperative appearance after orbital decompression surgery for thyroid eye disease[J]. Comput Biol Med 118:103628
Zhang L, Lu L, Nogues I et al (2017) DeepPap: deep convolutional networks for cervical cell classification[J]. IEEE J Biomed Health Inform 21(6):1633–1643
Zhao L, Yin J, Yuan L et al (2017) An efficient abnormal cervical cell detection system based on multi-instance extreme learning machine[C]. Ninth International Conference on Digital Image Processing (ICDIP 2017). Int Soc Opt Photon 10420:104203U
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jia, D., Zhou, J. & Zhang, C. Detection of cervical cells based on improved SSD network. Multimed Tools Appl 81, 13371–13387 (2022). https://doi.org/10.1007/s11042-021-11015-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11015-7