Abstract
Background
Acute myeloid leukemia (AML) is a type of blood cancer that affects the bone marrow and blood cells. AML is characterized by the rapid growth and accumulation of abnormal white blood cells, known as myeloblasts, which interfere with the production of normal blood cells.
Aims
The main aim was to determine the relationship between these genetic alterations and the clinico-haematological parameters and prognostic factors with therapy for Iraqi patients with AML.
Methods
We used Sanger Sequencing to detect the mutations in 76 AML patients. Clinical data of AML patients were retrospectively analysed to compare the prognosis of each gene mutation group.
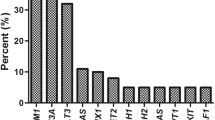
Results
Somatic mutations were identified in 47.4% of the enrolled patients in a core set of pathogenic genes, including FLT3 (18 patients, 23.7%), DNMT3A (14, 18.4%), NPM1 (11, 14.5%) and TP53 (5, 6.8%). As multiple mutations frequently coexisted in the same patient, we classified patients into 10 further groups. Two novel mutations were detected in FLT3-ITD, with new accession numbers deposited into NCBI GenBank (OP807465 and OP807466). These two novel mutations were computationally analysed and predicted as disease-causing mutations. We found significant differences between patients with and without the detected mutations in disease progression after induction therapy (remission, failure and death; pv = < 0.001) and statistically significant differences were reported in total leukocyte count (pv = < 0.0001).
Conclusion
These genes are among the most frequently mutated genes in AML patients. Understanding the molecular and clinical significance of these mutations is important for guiding treatment decisions and predicting patient outcomes.







Similar content being viewed by others
Data Availability
The datasets regarding FLT3-ITD gene generated during the study were deposited into the NCBI GenBank Banklt database and can be accessed with the accession numbers OP807465 and OP807466 (https://www.ncbi.nlm.nih.gov/nuccore/OP807465/https://www.ncbi.nlm.nih.gov/nuccore/OP807466). Furthermore, the data related to DNMT3A gene in our study released as: (https://www.ncbi.nlm.nih.gov/nuccore/ON881282.1) (https://www.ncbi.nlm.nih.gov/nuccore/ON881281.1) and (https://www.ncbi.nlm.nih.gov/nuccore/ON881280.1).
Code Availability
Not Applicable.
References
Arber DA et al (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391–2405
Tang S et al (2017) FLT3-ITD with DNMT3A R882 double mutation is a poor prognostic factor in chinese patients with acute myeloid leukemia after chemotherapy or allogeneic hematopoietic stem cell transplantation. Int J Hematol 106:552–561
Loghavi S et al (2014) Clinical features of de Novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J Hematol Oncol 7:1–10
Papaemmanuil E et al (2016) Genomic classification and prognosis in Acute myeloid leukemia. N Engl J Med 374:2209–2221
Volpe G et al (2015) Regulation of the flt3 gene in haematopoietic stem and early progenitor cells. PLoS ONE 10:1–18
Schlenk RF et al (2014) Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood 124:3441–3449
Verson CA (2020) Profiling FLT3 mutations in mexican Acute Myeloid Leukemia Pediatric Patients: impact on overall survival. 8,
Bhattacharyya J et al (2018) Prevalence and clinical significance of FLT3 and NPM1 mutations in Acute myeloid leukaemia patients of Assam, India. Indian J Hematol Blood Transfus 34:32–42
George B, Kantarjian H, Baran N, Krocker JD, Rios A (2021) Tp53 in acute myeloid leukemia: molecular aspects and patterns of mutation. Int J Mol Sci 22,
Huang X et al (2013) A novel PTEN/Mutant p53/ c-Myc/Bcl-XL axis mediates context-dependent oncogenic effects of PTEN with implications for cancer prognosis and therapy. Neoplasia (United States) 15:952–965
Hou H et al (2015) TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. https://doi.org/10.1038/bcj.2015.59
DiNardo C, Lachowiez C (2019) Acute myeloid leukemia: from mutation profiling to treatment decisions. Curr Hematol Malig Rep 14:386–394
Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia (2013) N Engl J Med 368:2059–2074
Bullinger L, Konstanze D (2022) Genomics of Acute Myeloid Leukemia Diagnosis and Pathways 35,
Lazenby M et al (2014) The prognostic relevance of flt3 and npm1 mutations on older patients treated intensively or non-intensively: a study of 1312 patients in the UK NCRI AML16 trial. Leukemia 28:1953–1959
Döhner H et al (2022) Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 140:1345–1377
Stein EM et al (2021) Safety and Efficacy of Menin Inhibition in patients (pts) with MLL-Rearranged and NPM1 mutant Acute Leukemia: a phase (ph) 1, first-in-human study of SNDX-5613 (AUGMENT 101). Blood 138:699–699
Uy GL et al (2021) Flotetuzumab as salvage immunotherapy for refractory acute myeloid leukemia. Blood 137:751–762
Untergasser A et al (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35:71–74
Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov K (2017) I. FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319
Manachai N et al (2022) Activating Mutation in the Receptor Tyrosine Kinase FLT3 with Clinicopathological Relevance in Canine Mast Cell Tumors. Vet. Med. Int (2022)
Park DJ et al (2020) Characteristics of DNMT3A mutations in acute myeloid leukemia. 55,
Gamaleldin MA, Imbaby SA, E. (2021) T. The role of tumor necrosis factor receptor superfamily member 4 (TNFRSF4) gene expression in diagnosis and prognosis of acute myeloid leukemia. Mol Biol Rep 48:6831–6843
Kadia TM et al (2016) TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomes. Cancer 122:3484–3491
Ali AM, Salih GF (2022) Identification of three novel DNMT3A mutations with compromising methylation capacity in human acute myeloid leukaemia. Mol Biol Rep. https://doi.org/10.1007/s11033-022-07977-y
Ali KM et al Clinical outcomes and phylogenetic analysis in reflection with three predominant clades of SARS-CoV-2 variants. 0–2 https://doi.org/10.1111/eci.14004
ProtParam tool. https://web.expasy.org/protparam/
Yunus NM et al (2015) Characterisation and clinical significance of FLT3-ITD and non-ITD in Acute myeloid leukaemia patients in Kelantan. Northeast Peninsular Malaysia. https://doi.org/10.7314/APJCP.2015.16.12.4869
Laskowski RA, Thornton JM, VarSite (2020) Disease variants and protein structure. 111–119. https://doi.org/10.1002/pro.3746
Falini B et al (2006) Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood 108:1999–2005
Othman GO, Mohammad NS, Saeed CH (2021) Molecular study of nucleophosmin 1 (NPM1) gene in acute myeloid leukemia in kurdish population. 21:687–692
Yanada M et al (2016) TP53 mutations in older adults with acute myeloid leukemia. Int J Hematol 103:429–435
Dutta S et al (2020) Functional classification of tp53 mutations in acute myeloid leukemia. Cancers (Basel) 12:1–16
Article O (2020) Impact of the variant allele frequency of ASXL1, DNMT3A, JAK2, TET2, TP53, and NPM1 on the Outcomes of patients with newly diagnosed Acute myeloid leukemia. 765–774. https://doi.org/10.1002/cncr.32566
Hamed HR, Al-jumaily RMK, Kadhom AE (2021) Characterization of NPM1 and FLT3-ITD mutations in iraqi patients with AML. 21:133–139
Kiyoi BH et al (1999) Prognostic implication of FLT3 and N- RAS gene mutations in Acute myeloid leukemia. 93:3074–3080
Sazawal S et al (2017) NPM1 and FLT3 mutations in acute myeloid leukemia with normal karyotype. Indian perspective. https://doi.org/10.4103/IJPM.IJPM
Kottaridis PD et al (2001) The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United King. Blood 98:1752–1759
Rashid PMA, Salih GF (2022) Molecular and computational analysis of spike protein of newly emerged omicron variant in comparison to the delta variant of SARS – CoV – 2 in Iraq. Mol Biol Rep. https://doi.org/10.1007/s11033-022-07545-4
Cheng J, Qu L, Wang J, Cheng L, Wang Y (2018) High expression of FLT3 is a risk factor in leukemia. Mol Med Rep 17:2885–2892
Naseem S et al (2021) NPM1 and FLT3-ITD / TKD gene mutations in. 15:15–26
Hindley A, Catherwood MA, McMullin MF, Mills K (2021) I. significance of npm1 gene mutations in aml. Int J Mol Sci 22,
Sasaki K et al (2020) Impact of the variant allele frequency of ASXL1, DNMT3A, JAK2, TET2, TP53, and NPM1 on the outcomes of patients with newly diagnosed acute myeloid leukemia. Cancer 126:765–774
Döhner H et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129:424–447
Gary Gilliland D, Griffin JD (2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100:1532–1542
Hou HA et al (2012) DNMT3A mutations in acute myeloid leukemia: Stability during disease evolution and clinical implications. Blood 119:559–568
Naoe T, Kiyoi H (2013) Gene mutations of acute myeloid leukemia in the genome era. 165–174. https://doi.org/10.1007/s12185-013-1257-4
Elrhman HAEA, El-Meligui YM, Elalawi SM (2021) Prognostic impact of concurrent DNMT3A, FLT3 and NPM1 gene mutations in Acute myeloid leukemia patients. Clin Lymphoma Myeloma Leuk 21:e960–e969
Easwar A, Siddon AJ (2021) Genetic landscape of myeloproliferative neoplasms with an emphasis on molecular diagnostic laboratory testing. Life 11:1–22
Bumbea H et al (2022) Platelet Defects in Acute Myeloid Leukemia — Potential for Hemorrhagic Events.
Bond J et al (2019) T-cell acute lymphoblastic leukemia 104:1617–1625
Stone RM et al (2017) Midostaurin plus chemotherapy for Acute myeloid leukemia with a FLT3 mutation. N Engl J Med 377:454–464
Thiede C et al (2020) Impact of NPM1 / FLT3 -ITD genotypes de fi ned by the 2017 european LeukemiaNet in patients with acute myeloid leukemia. 135:371–380
Barranco R, Fossati F, Candosin S, Orcioni GF, Ventura F (2016) Unexpected sudden death due to acute myeloid leukemia subtype M5: a case report and review of the literature. 207–211. https://doi.org/10.4323/rjlm.2016.207
Kiani BH et al (2020) Artemisinin and its derivatives: a promising cancer therapy. Mol Biol Rep 47:6321–6336
Acknowledgements
Not applicable.
Funding
Not Applicable.
Author information
Authors and Affiliations
Contributions
AMA and GFA were involved in the conception of the study idea and the experimental design. AMA conducted the laboratory work, performed data analysis, and prepared the manuscript. GFA provided supervision and guidance throughout the process. Both authors participated in discussions regarding the results and contributed to the finalization of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethical aspects of this study were given utmost importance and followed the established guidelines and regulations. Prior to enrollment, all patients provided verbal consent for the collection of blood samples. The study protocol was reviewed and approved by the Ministry of Health’s health directorate, with the approval obtained on August 23, 2021 (protocol NO. 9811). The research investigations were conducted at the University of Sulaimani, College of Science, in accordance with the applicable rules and regulations. The university granted approval for the study (No. 1917/259 on 01/08/2021), ensuring that ethical standards were upheld and necessary protocols were followed throughout the study. The study was conducted in compliance with the research ethical guidelines set by the Ministry of Health. The ethical standards outlined in the Declaration of Helsinki, which governs medical research involving human subjects, were strictly followed. Participants, including the parents of patients under the age of 18, provided both verbal and written consent after being provided with the necessary information and having the opportunity to review the Declaration of Helsinki.
Consent to participant
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ali, A.M., Salih, G.F. Molecular and clinical significance of FLT3, NPM1, DNMT3A and TP53 mutations in acute myeloid leukemia patients. Mol Biol Rep 50, 8035–8048 (2023). https://doi.org/10.1007/s11033-023-08680-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08680-2