Abstract
Background
Previous studies have revealed a reduction in the genetic diversity of P. trituberculatus in Bohai Sea. However, because swimming crabs have been released into this area for some time, it is unclear whether the release of cultured populations from the national breeding farms of swimming crabs in Bohai Bay have affected the population genetics of wild populations of P. trituberculatus.
Methods and results
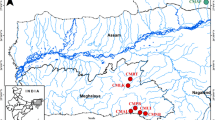
In this study, the genetic diversity and population structure of 120 P. trituberculatus specimens in Bohai Bay were investigated using six microsatellite loci, including one wild population and one cultivated population. A total of 132 alleles were identified for all loci. The mean expected (He) and observed heterozygosity (Ho) were 0.8185 and 0.7759, respectively, thus indicating high levels of genetic diversity for these two populations. Molecular variance analysis (AMOVA) (FST = 0.0180), genetic distance (D = 0.1168) and similarity (S = 0.8898) indicated that these two populations could not be distinguished genetically. Structural analysis, phylogenetic tree construction, and principal component analysis, showed no significant distinction between the wild and cultivated populations. Finally, we investigated genetic exchange between the two populations by analyzing migration rate (M) and gene flow (Nm), thus demonstrating significant flow of genetic information.
Conclusions
These findings contribute information for further breeding schemes for P. trituberculatus, evaluation of its genetic potential and programs for the protection of wild resources.



Similar content being viewed by others
References
Fishery Bureau, Ministry of Agriculture PRC (FBMAPRC) (2021) China fisheries yearbook 2021. Chinese Agriculture Express, pp 22–38
Min JJ, Ye RH, Zhang GF, Zheng RQ (2015) Microsatellite analysis of genetic diversity and population structure of freshwater mussel (Lamprotula leai). Zool Res 36:34–40
Liu S, Xue SX, Sun JS (2008) Genetic diversity of Portunus triuberbuculatus revealed by AFLP analysis. Oceanol Limnol Sin 39:152–156
Chi DL, Yan BL, Shen SD, Gao H (2010) RAPD analysis between color-different crab individuals of Portunus trituberculatus. Mar Sci Bull (Chin Ed) 12:47–54
Guo EM, Liu Y, Cui ZX, Li XL, Cheng YX, Wu XG (2012) Genetic variation and population structure of swimming crab (Portunus trituberculatus) inferred from mitochondrial control region. Mol Biol Rep 39:1453–1463
Lu CY, Kuang YY, Zheng XH, Li C, Sun XW (2019) Advances of molecular marker-assisted breeding for aquatic species. J Fish China 43:38–55
Guo EM, Cui ZX, Wu DH, Hui M, Liu Y, Wang HX (2013) Genetic structure and diversity of Portunus trituberculatus in Chinese population revealed by microsatellite markers. Biochem Syst Ecol 50:313–321
Chistiakov DA, Hellemans B, Volckaert FAM (2006) Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255:1–29
Verma H, Borah JL, Sarma RN (2019) Variability assessment for root and drought tolerance traits and genetic diversity analysis of rice germplasm using SSR markers. Sci Rep 9:16513
Li XP, Li P, Li J, Gao BQ (2011) Genetic diversity among five wild populations of Portunus trituberculatus. J Fish Sci China 18:1327–1334
Li XP, Liu P, Song XF (2011) Construction on enriched microsatellite library and characterization of microsatellite markers from swimming crab. J Fish Sci China 18:194–201
Liu YG, Guo YH, Hao J, Liu LX (2012) Genetic diversity of swimming crab (Portunus trituberculatus) populations from Shandong peninsula as assessed by microsatellite markers. Biochem Syst Ecol 41:91–97
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Yeh FC, Yang RC, Boyle T (1999) POPGENE version1.32. Microsoft Window-base Software for Population Genetic Analysis, A Quick User’s Guide. University of Alberta, Center for International Forestry Research, Alberta, Canada
Rousset F (2008) GENEPOP’007: a complete reimplementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0), an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Earl DA, Vonholdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Beerli P, Palczewski M (2010) Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185:313–326
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Zhai SN, Comes HP, Nakamura K, Yan HF, Qiu YX, Silman MR (2012) Late Pleistocene lineage divergence among populations of Neolitsea sericea (Lauraceae) across a deep sea-barrier in the Ryukyu Islands. J Biogeogr 39:1347–1360
Qin Y, Shi G, Sun Y (2013) Evaluation of genetic diversity in Pampus argenteus using SSR markers. Genet Mol Res 12:5833–5841
Xu QH, Liu R (2011) Development and characterization of microsatellite markers for genetic analysis of the swimming crab, Portunus trituberculatus. Biochem Genet 49:202–212
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Li PF, Liu P, Li J, Dai FY, He Y (2007) Biochemical genetic analysis of Portunus trituberculatus. Mar Fish Res 28:90–96
Feng BB, Li JL, Niu DH, Chen L, Zhou ZQ, Zheng YF, Zheng KH (2008) Sequence analysis of mitochondrial putative control region gene fragments of wild Portunus trituberculatus in four sea regions in China. J Shanghai Ocean Univ 17:134–139
Hui M, Shi GH, Sha ZL, Liu Y, Cui ZX (2018) Genetic population structure in the swimming crab, Portunus trituberculatus and its implications for fishery management. J Mar Biol Assoc UK 99:891–899
Wright S (1972) Evolution and the genetics of populations. A treatise in four volumes. Volume 4. Variability within and among natural populations. J Biosocial Sci 4:253–256
Wright S (1931) Evolution in Mendelian population. Genetics 16:91–159
Li PF, Xu KD, Zhou HX (2015) Genetic diversity of Portunus trituberculatus among the cultured and wild populations in the offing of Zhejiang using microsatellite markers. J Zhejiang Ocean Univ Nat Sci 34:537–542
Rousset F, Raymond M (1995) Testing heterozygote excess and deficiency. Genetics 140:1413–1419
Wallace B (1988) Molecular evolutionary genetics. J Hered 79:139
Chai CJ, Esa YB, Ismail MFS, Kamarudin MS (2017) Population structure of the blue swimmer crab Portunus pelagicus in coastal areas of Malaysia inferred from microsatellites. Zool Stud 56:26
Liu Q, Cui F, Hu P, Yi GT, Ge YW, Liu WL, Yan HW, Wang LS, Liu HY, Song J, Jiang YS, Zhang L, Tu Z (2018) Using of microsatellite DNA profiling to identify hatchery-reared seed and assess potential genetic risks associated with large-scale release of swimming crab Portunus trituberculatus in Panjin, China. Fish Res 207:187–196
Liu S, Sun JS, Hurtado LA (2013) Genetic differentiation of Portunus trituberculatus, the world’s largest crab fishery, among its three main fishing areas. Fish Res 148:38–46
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn/) for the expert linguistic services provided.
Funding
This work was supported by the Hebei Natural Science Foundation (C2016201249), Science and Technology Innovation Project of Modern Seed Industry (21326307D), Institute of Life Science and Green Development (Hebei University) and Innovation Center for Bioengineering and Biotechnology of Hebei Province.
Author information
Authors and Affiliations
Contributions
BD: Conceptualization, Methodology, Software, Writing original draft. WL: Data curation, Formal analysis. SL: Data curation, Formal analysis. YY: Investigation, Formal analysis. YG: Validation, Methodology. SM and XK: Supervision, Project administration, Conceptualization, Writing-Review & Editing. ZL and XJ: Resources.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All animal experiments were compliant with the local approved protocols of the Administration Office Committee for Laboratory Animals, and aimed to minimize animal suffering.
Informed consent
All authors informed consent.
Consent to publish
All authors approved the version to be published.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Duan, B., Liu, W., Li, S. et al. Microsatellite analysis of genetic diversity in wild and cultivated Portunus trituberculatus in Bohai Bay. Mol Biol Rep 49, 2543–2551 (2022). https://doi.org/10.1007/s11033-021-07054-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-07054-w