Abstract
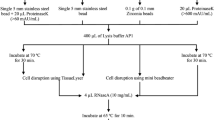
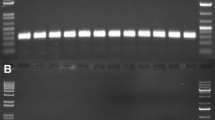
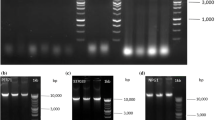
Effective isolation of high-quality genomic DNA is one of the essential steps in molecular biology, biochemistry, and genetic studies. Here we describe a simplified procedure based on repeated freeze–thawing cycles to isolate genomic DNA from different organisms of microbes (Burkholderia pyrrocinia JK-SH007, Bacillus pumilus HRl0, Botrytis cinerea) and nematodes (Bursaphelenchus xylophilus). The DNA extraction buffer includes 10% of CTAB; 4% of NaCl (W/V); 20 mM of ethylenediamine tetraacetic acid; 100 mM of Tris-HCl, pH 8.0 and 1% of polyvinylpyrrolidone. The released DNA was purified from the mixture using a phenol/chloroform mixture and precipitated in 70% ethanol to remove proteins, carbohydrates, phenols, RNA, etc. Our method is a reproducible, simple, and rapid technique for routine DNA extractions from various microorganisms and nematodes. Furthermore, the low cost of this method could be an economic benefit to large-scale studies.



Similar content being viewed by others
Data availability
The data used to support the findings of this study are included in the article.
References
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3(2):208–218. https://doi.org/10.1016/S0022-2836(61)80047-8
Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich CM (2009) Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip 9:2811–2817. https://doi.org/10.1039/B905065P
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. Springer, Dordrecht, pp 183–190
Kuo TT, Chao YS, Lin YH, Lin BY, Liu LF, Feng TY (1987) Integration of the DNA of filamentous bacteriophage Cflt into the chromosomal DNA of its host. J Virol 61(1):60–65
Mahuku GS (2004) A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol Biol Rep 22(1):71–81. https://doi.org/10.1007/BF02773351
Lachica RVF, Hoeprich PD, Genigeorgis C (1971) Nuclease production and lysostaphin susceptibility of Staphylococcus aureus and other catalase-positive cocci. Appl Environ Microbiol 21(5):823–826
Luo J, Yang J, He H, Jin T, Zhou L, Wang M, Zhou M (2013) A new electrochemically active bacterium phylogenetically related to Tolumonas osonensis and power performance in MFCs. Bioresour Technol 139:141–148. https://doi.org/10.1016/j.biortech.2013.04.031
Imai T, Ohta K, Kigawa H, Kanoh H, Taniguchi T, Tobari J (1994) Preparation of high-molecular-weight DNA: application to mycobacterial cells. Anal Biochem 222(2):479–482. https://doi.org/10.1006/abio.1994.1520
Li JT, Yang J, Chen D, Zhang X, Tang Z (2007) An optimized mini-preparation method to obtain high-quality genomic DNA from mature leaves of sunflower. Genet Mol Res 6(4):1064–1071
Johnson BH, Hecht MH (1994) Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Nat Biotechnol 12(12):1357–1360. https://doi.org/10.1038/nbt1294-1357
Silva GAD, Bernardi TL, Schaker PDC, Menegotto M, Valente P (2012) Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz Arch Biol Technol 55(2):319–327. https://doi.org/10.1590/S15168913201200020-0020
Ren JH, Ye JR, Liu H, Xu XL, Wu XQ (2011) Isolation and characterization of a new Burkholderia pyrrocinia strain JK-SH007 as a potential biocontrol agent. World J Microbiol Biotechnol 27(9):2203–2215. https://doi.org/10.1007/s11274-011-0686-6
Sheng J, Wu X, Hou L (2014) Isolating and identifying mycorrhiza helper bacteria from the rhizosphere soil of Pinus thunbergi inoculated with Rhizipogen luteous. J Northeast For Univ 42(5):110–114. https://doi.org/10.13759/j.cnki.dlxb.20140522.036
Qu HY, Tan JJ, Ye JR, Hao DJ, Huai YJ (2009) Effects of different fungus on the reproduction and virulence of Bursaphelenchus xylophilus. J Nanjing For Univ 33(6):57–59. https://doi.org/10.3969/j.issn.1000-2006.2009.06.013
Chen YH, Ye JR, Wei CJ, Yang XM (2002) Effects of pine wood nematode infection on metabolism of active oxygen in japanese black pine and slash pine seedlings. J Nanjing For Univ 26(4):19–22. https://doi.org/10.3969/j.issn.1000-2006.2002.04.005
Mathew B, Ugboko H, De N (2015) Prevalence of multidrug resistant Salmonella enterica serovar Typhi in Kaduna Metropolis, Kaduna, Nigeria. Int J Curr Microbiol Appl Sci 4(9):323–335
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. PNAS 81:8014–8018. https://doi.org/10.1073/pnas.81.24.8014
Rang J, Li L, Tang Q, Qi Yang, He L, Ding X, Xia L (2015) Comparative study of bacterial DNA extraction methods for the third generation sequencing technology. J Nat Sci Hunan Norm Univ 38(6):14–20. https://doi.org/10.7612/j.issn.1000-2537.2015.06.003
Carillo S, Silipo A, Perino V, Lanzetta R, Parrilli M, Molinaro A (2009) The structure of the O-specific polysaccharide from the lipopolysaccharide of Burkholderia anthina. Carbohydr Res 344(13):1697–1700. https://doi.org/10.1016/j.carres.2009.05.013
Wu F, Huang D, Huang X, Xin Z, Cheng W (2009) Comparing Study on several Methods for DNA extraction from endophytic fungi. Chin Agric Sci Bull 25:62–64
Guo H, Chao Y, Guo W, Bao Q, Sun W, Qian A (2011) Comparative study of microbial genome dna extraction methods from the gastrointestinal tract of murine embryos. Biotechnology 21(4):37–40. https://doi.org/10.3969/j.issn.1004-311X.2011.04.096
Chen WP, Kuo TT (1993) A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res 21(9):2260. https://doi.org/10.1093/nar/21.9.2260
Zhu H, Qu F, Zhu LH (1993) Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res 21(22):5279–5280. https://doi.org/10.1093/nar/21.22.5279
Acknowledgements
This work was financially supported by the Jiangsu Agricultural Science and Technology Independent Innovation Fund (CX16-1005) and the Chinese State Forestry Administration Special Research Program for Forestry Sectors Beneficial to the Public (No. 201304404). This work was also financially supported by the Shanghai Science and Technology Agriculture Key Project (2014:5-6) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, F., Ye, J., Chio, C. et al. A simplified quick microbial genomic DNA extraction via freeze–thawing cycles. Mol Biol Rep 47, 703–709 (2020). https://doi.org/10.1007/s11033-019-05176-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05176-w