Abstract
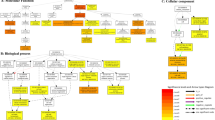
Plants have continually confrontation with different abiotic stresses, including salt, low temperature, drought or hormone stress. The plants acclimate to the environmental stresses relating with the falls of the molecular mesh including the stress signal receiver, signal transcriptional regulation and the expression of functional and structure genes. Using the RNA-seq, we carried out a transcriptional analysis under cold treatment for investigating a profound comprehension of the signal network and molecular metabolisms reaction included in abiotic stress reaction for Lilium lancifolium. Our study identified 18,722 unigenes had demonstrated the resemblance to the known exact proteins in the Swiss-Prot protein database and classified them by Gene ontology into three primary kinds: cellular component, biological process, and molecular function, and then 15,898 unigenes aligned to existing sequences in the KEGG databases. Based on the transcriptome results of cold stress, more stress-related genes were identified and analyzed of their expressions in other abiotic stress treatments as 37 °C, ABA, JA and Na. Meanwhile, bioinformatics qRT-PCR analyses of stress genes as LlDREB1, LlAP2, LlNAC1, LlHOT, LlR2R3-MYB and LlCDPK revealed that novel candidate genes encoding ethylene responsive transporters and serine/threonine receptor-like kinases, which contributed to speculate the signal regulation pathway during the abiotic stresses; engineering genes could also boost the tolerance to stress, as protected and maintained the function and structure of cellular components. Our research conjectured the abiotic stress signal transduction pathway and identified the expected key ingredients regulating the stress tolerance in Lilium lancifolium, which would enable the in-depth molecular exploration of stress-tolerance mechanisms in lily.










Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- DREB:
-
Dehydration responsive element binding protein
- EREBP:
-
Ethylene-responsive element binding protein
- MYB:
-
Myeloblastosis
- NAC:
-
NAM, ATAF1-2, CUC2
- TF:
-
Transcription factor
References
Bray EA, Bailey-Serres J: Responses to abiotic stresses (chapter 22). In: W Gruissem, B Buchannan, R Jones (2000) Responses to abiotic stresses. American Society of Plant Physiologists, Rockville, pp 1158–1249
Kaoru U, Yukio K, Motoaki S, Kazuo S (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13:132–138
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14:194–199
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heatshock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Park EJ, Jeknic Z, Sakamoto A, Denoma J, Yuwansiri R, Murata N, Chen TH (2004) Genetic engineering of glycinebetaine synthesis in tomato protects seeds, plants, and flowers from chilling damage. Plant J 40:474–487
Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13:146–150
Rontein D, Basset G, Hanson AD (2002) Metabolic engineering of osmoprotectant accumulation in plants. Metab Eng 4(49–56):9
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Zhang JZ, Creelman RA, Zhu JK (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Wang JM, Yang Y, Liu XH (2014) Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genom 15:203
Tsirigos A, Haiminen N, Bilal E, Utro F (2012) A computational platform for developing high-throughput analytics in genomics. Bioinformatics 28:282–283
Kakumanu A, Ambavaram MMR, Klumas C, Krishnan A, Batlang U (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160:846–867
O’Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu J, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D (2013) An RNA-Seq transcriptome analysis of Pi deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161:705–724
Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, Chen L (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20(2):646–654
Raherison E, Rigault P, Caron S, Poulin P, Boyle B, Verta JP, Giguere I, Bomal C, Bohlmann J, MacKay J (2012) Transcriptome profiling in conifers and the PiceaGen Express database show patterns of diversification within gene families and interspecific conservation in vascular gene expression. BMC Genom 13:434–449
Qiu Q, Ma T, Hu Q, Liu B, Wu Y, Zhou H, Wang Q, Wang J, Liu J (2011) Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol 31(4):452–461
Xu DL, Long H, Liang JJ, Zhang J, Chen X, Li JL, Pan ZF, Deng GB, Yu MQ (2012) De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genom 13:133–137
Cong HQ, Li ZY, Xu L (2013) Characterizing developmental and inducible differentiation between juvenile and adult plants of Aechmea fasciata treated with ethylene by transcriptomic analysis. Plant Growth Regul 69:247–257
Trick M, Long Y, Meng J, Bancroft I (2009) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J 7(4):334–346
Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T, Brutnell TP (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42(12):1060–1067
Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y (2012) De novo assembly and Characterisation of the Transcriptome during seed development, and generation of genic-SSR markers in Peanut (Arachis hypogaea L.). BMC Genom 13:90
Gahlan P, Singh HR, Shankar R, Sharma N, Kumari A, Chawla V, Ahuja PS, Kumar S (2012) De novo sequencing and characterization of Picrorhiza kurrooa transcriptome at two temperatures showed major transcriptome adjustments. BMC Genom 13:126
Chen JH, Song YP (2013) Genome-Wide analysis of gene expression in response to drought stress in Populus simomnii. Plant Mol Biol Rep 31:946–996
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Qiu ZB, Wan LC (2013) The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol 199:627–870
Xin Z (2001) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell Environ 23:893–902
Shulaev V, Cortes D, Miller G, Mittler R (2008) Metabolomics for plant stress response. Physiol Plant 132:199–208
Silva JMd, Arrabac MC (2004) Contributions of soluble carbohydrates to the osmotic adjustment in the C4 grass Setaria sphacelata: a comparison between rapidly and slowly imposed water stresses. J Plant Physiol 161:551–555
Capell T, Christou P (2004) Progress in plant metabolic engineering. Curr Opin Biotechnol 15:148–154
Konstantinova T, Parvanova D, Atanassov A, Djilianov D (2002) Freezing tolerant tobacco, transformed to accumulate osmoprotectants. Plant Sci 163:157–164
Cooper GM, Hausman RE (2012) The cell: a molecular approach. Beijing Sci Publ 15:603–605. ISBN 978-7-03-031324-9
Donald WH, Rebecca B (1996) Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science 273:956–959
Acknowledgments
This work was supported by the ‘863’ research program (Grant No. 2011AA10020804), China National Natural Science Foundation (Grant No. 31071815 and No. 31272204), and the D. Programs Foundation of the Ministry of Education of China (Grant No. 20110014110006).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2014_3725_MOESM1_ESM.doc
Availability of supporting data: The supporting data set could be available in the NCBI GenBank repository (http://www.ncbi.nlm.nih.gov/genbank/), the ID numbers are KJ489026, KJ489025, KJ467617, KJ489024, KJ467624, KJ467618, KJ467623, KJ467622, KJ467620, KJ467621. Additional file 1: Table S1. Expressions of different genes using the Heat-map. Heat-map of 102 differentially expressed genes involved in the cold response and acclimation of Lilium lancifolium. (DOC 135 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Wang, Q., Yang, Y. et al. De novo assembly and characterization of stress transcriptome and regulatory networks under temperature, salt and hormone stresses in Lilium lancifolium . Mol Biol Rep 41, 8231–8245 (2014). https://doi.org/10.1007/s11033-014-3725-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3725-1