Abstract
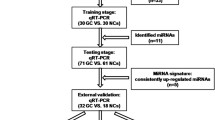
Gastric cancer is the fourth most prevalent malignancy worldwide and remains the second most common cause of cancer-related death globally. Understanding the molecular structure of gastric carcinogenesis might identify new diagnostic and therapeutic strategies for this disease. Thus, early detection of gastric cancer is a key measure to reduce the mortality and improve the prognosis of gastric cancer. There have recently been several reports that microRNAs (miRNAs) circulate in highly stable, cell-free forms in blood. Because serum and plasma miRNAs are relatively easy to access, circulating miRNAs also have great potential to serve as non-invasive biomarkers. Although a number of miRNAs associated with gastric cancer have been identified, the underlying mechanism of these miRNAs in tumorigenesis and tumor progression remains to be investigated. The purpose of this study is to identify the potential of serum miRNAs as biomarkers for early detection of gastric cancer patients. RNA was isolated using the High Pure miRNA Isolation Kit (Roche) following the manufacturer’s protocol. cDNA and preamplification protocols were obtained from the isolated plasma miRNAs. The BioMark™ 96.96 Dynamic Array (Fluidigm Corporation) for real-time qPCR was used to simultaneously quantite the expression of 740 miRNAs. All statistical analyses were performed using the Biogazelle’s qbase PLUS 2.0 software. In this study, among 740 miRNAs that we analyzed only miR-195-5p was significantly (p < 0.05, fold changes = 13, 3) down-regulated in gastric cancer patients compared with control. We demonstrated that miR-195-5p is a novel tumor suppressor miRNA and may contribute to gastric carcinogenesis. The miRNA expression profile described in this study should contribute to future studies on the role of miRNAs in gastric cancer.

Similar content being viewed by others
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55:74–108
Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D et al (2010) Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol 45(1):17–23
Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T et al (2010) Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102(7):1174–1179
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101(10):2087–2092. doi:10.1111/j.1349-7006.2010.01650.x
Olsen PH, Ambros V (1999) The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 216(2):671–680
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J et al (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433(7027):769–773
Lu M, Zhang Q, Deng M, Miao J, Guo Y et al (2008) An Analysis of human microRNA and disease associations. Plos One 3(10):e3420. doi:10.1371/journal.pone.0003420
Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY et al (2012) Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. Plos One 7(3):e33608
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302(1):1–12
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W et al (2008) miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Investig 88(12):1358–1366
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K et al (2008) Detection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141:672–675
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI (2008) Mature miR- 184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14:2588–2592
Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J et al (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58:1375–1381
Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE (2009) The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol 112:55–59
Ma YY, Tao HQ (2012) Microribonucleic acids and gastric cancer. Cancer Sci 103(4):620–625
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L et al (2009) MicroRNA profiling of human gastric cancer. Mol Med Rep 2(6):963–970
Chen L, Yang Q, Kong WQ, Liu T, Liu M, Li X et al (2012) MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life 64(7):628–635
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC et al (2012) Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Gene Chromos Cancer 51(4):394–401
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C et al (2011) A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 47(5):784–791
Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S et al (2010) Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 11(2):136–146
Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y et al (2009) Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta 400(1–2):97–102
Acknowledgments
This work which is the part of “Establishment of Molecular Substructure Laboratory for Early Diagnosis of Cancer (TR62-09-03)” project was supported by Grants from Çukurova Development Agency (ÇKA), Mersin, Turkey.
Conflict of interest
No conflict of interest is declared by the author(s).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gorur, A., Balci Fidanci, S., Dogruer Unal, N. et al. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep 40, 2091–2096 (2013). https://doi.org/10.1007/s11033-012-2267-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2267-7