Abstract
A full-length cDNA of 1,728 nt, called MsLEA3-1, was cloned from alfalfa by rapid amplification of cDNA ends from an expressed sequence tag homologous to soybean pGmPM10 (accession No. AAA91965.1). MsLEA3-1, encodes a deduced protein of 436 amino acids, a calculated molecular weight of 47.0 kDa, a theoretical isoelectric point of 5.18, and closest homology with late embryogenesis abundant proteins in soybean. Sequence homology suggested a signal peptide in the N terminus, and subcellular localization with GFP revealed that MsLEA3-1 was localized preferentially to the nucleolus. The transcript titre of MsLEA3-1 was strongly enriched in leaves compared with roots and stems of mature alfalfa plants. Gene expression of MsLEA3-1 was strongly induced when seedlings were treated with NaCl and ABA. Expression of the MsLEA3-1 transgenic was detected in transgenic tobacco. Malondialdehyde content and, electrical conductivity content were reduced and electrical conductivity and proline content were increased in transgenic tobacco compared with non-transgenic tobacco under salt stress. The results showed that accumulation of the MsLEA3-1 protein in the vegetative tissues of transgenic plants enhanced their tolerance to salt stress. These results demonstrate a role for the MsLEA3-1 protein in stress protection and suggest the potential of the MsLEA3-1 gene for genetic engineering of salt tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the start of the 20th century, the explosive increase in world population and continuous extreme environmental conditions have disrupted the overall homeostasis of plants, posing serious threats to global agricultural production and adverse agricultural environments [1]. Despite focused efforts to improve major crop species by traditional breeding, success has been limited in developing tolerance to to abiotic stress. Some plants have developed physiological and biochemical strategies to adapt or tolerate extreme stress conditions on their own, and relevant genes can be isolated and exploited to develop tolerance in other plant species. Reduction of sodium ions in the cytoplasm and the accumulation of compatible, low molecular weight protective compounds (called osmolytes) have been suggested as two major mechanisms that underlie the adaptation or tolerance process [2].
In addition to metabolic changes, a large set of plant genes commonly called LEA (LATE EMBRYOGENESIS ABUNDANT) are transcriptionally activated and appear to lead to the accumulation of novel vegetative proteins in plants under osmotic stress [3]. LEA proteins are produced late during embryo development and constitute about 4% of the total cellular proteins. They were first characterized in cotton as a set of proteins that are highly accumulated in the embryos at the late stage of seed development. They appear to have widespread distribution among plant species [4]. These proteins are expressed in seeds as well as in drought stressed tissues where they play a major role as cellular protectants. LEA proteins are classified into seven subgroups, based on their amino acid sequence homology and specific motifs, which presumably undertake diverse functions during the periods of water deficit [5, 6]. Several groups of LEA protein genes, especially groups 2, 3 and 5, have been demonstrated to confer water-deficit and salt-stress tolerance.
Tolerance to abiotic stress is a complex trait that involves multiple physiological and biochemical mechanisms and regulation of numerous genes. Genetic engineering is believed to be an alternate, potentially important approach for creating new cultivars with better adaptation. Even single-gene transfers have led to the development of tolerant plants. For example, LE25, which belongs to Group 5 LEA protein from tomato (Lycopersicum esculentum L.), improved resistance to high salinity and freezing temperatures when expressed in Saccharomyces cereisiae [7]. Similarly, expression of the rice transcription factor, OsDREB1A, generated tolerance to drought, high salt and freezing stresses in transgenic Arabidopsis [8]. The hiC6 gene encoding a homologue of LEA enhances freezing tolerance of S. cerevisiae [9]. These examples focused on greenhouse evaluation of plant phenotypes, whereas documentation of phenotypes and expression of LEA genes is relatively less frequent in field plants. However, expression of HVA1, the Group 3 LEA protein from barley (Hordeum ulgare L.) conferred tolerance to soil water-deficit and salt stress in transgenic rice plants [10–12]. Thus, transgenetic manipulation of agriculturally important plants for enhanced abiotic stress tolerance seems to be a promising strategy.
Alfalfa (Medicago sativa L.) is a perennial legume forage, which is highly adaptable and has the largest planting area in the world. It has the characteristics of high forage yield, rich nutritional value and good palatability, and is named “the Queen of forages”. Since water-deficit and salinity are dominant restrictions for alfalfa. In this report, we present the molecular and functional characterization of MsLEA3-1, a new alfalfa gene that encodes a hydrophobic LEA protein. We also demonstrate that MsLEA3-1, which specifically expressed in alfalfa leaves and roots, responds to NaCl and ABA stresses and is localized preferentially to nucleolus. Furthermore, it offers tolerance to salt stress in transgenic Nicotiana plants expressing MsLEA3-1. This MsLEA-1 also has the potential to facilitate the generation of new stress-resistance alfalfa cultivars using gene transfer technology.
Materials and methods
Plant materials and growth conditions
Alfalfa (Medicago sativa cv. “Zhongmu NO.1”, bred by the Chinese Academy of Agricultural Sciences (CAAS) in 1997, was obtained from the Chinese Academy of Agricultural Sciences. Seeds were placed on MS agar media, then germinated in the dark at 25°C for 1 day. Alfalfa seeds were sown into soil and were grown with a photosynthetic flux of 400 μmol m−2 s−1, temperature 26–28°C, 16 h light (250 μE m−2 s−1)/8 h dark cycle, and 60–80% relative humidity. Seeds of Nicotiana benthamiana Domin conserved by CAAS were sterilized and sown aseptically on MS medium as above for alfalfa and supplemented with 100 mg l−1 myo-inositol, 3% (w/v) sucrose, 0.2% (w/v) phytagel and pH adjusted to 5.8. Nicotiana plants were grown with the same condition as for alfalfa.
Amplification of full-length cDNA
A 409 bp EST isolated from a salt-induced SSH cDNA library [13] of alfalfa was used as a template for rapid amplification of 3′ and 5′ cDNA ends using 5′GSP, 3′GSP, 5′GSPnest and universal primers (Table 1), a SMART™ RACE cDNA amplification kit (Clontech and an Advantage™ 2 PCR Enzyme kit (Clontech, Japan) following the manufacturer’s protocol. Total RNA was isolated from 10-day-old alfalfa seedlings by a TRIZOL reagent (Invitrogen, USA) according to manufacturer’s instructions. Amplicons were treated with DNase (Promega) and diluted 100-fold. RACE products were separated by electrophoresis on a 1% agarose gel stained with ethidium bromide, and extracted using a kit (Takara, Japan). The products were cloned into the pMD-19T vector (Takara, Japan) and then used to transform Escherichia coli DH5α. Recombinant sequenced plasmids were by the Huada Gene Company (Beijing, China) and the 5′- and 3′-ends linked together using an overlapping fragment of the LEA gene from soybean by DNAMAN software to form the full-length cDNA. Accordingly, a pair of primers, F-cDNA and R-cDNA with BamHI and EcoRI as restriction enzyme sites (Table 1), respectively, were designed and used to amplify the full-length cDNA from cv. Zhongmu No 1 seedlings stressed with salt using an Advantage 2 PCR Enzyme System (Clontech, Japan) according to the manufacturer’s instructions. The full-length amplicon was eluted, cloned, and sequenced as described above and designated MsLEA3-1 and deposited at the National Center for Biotechnology Information (NCBI) under accession number EU665182.
MsLEA3-1 sequence analysis
Homology searches were performed with BLAST and BLASTX algorithms to confirm sequence identity at NCBI (http://www.ncbi.nlm.nih.gov/) [14]. A search for open reading frames (ORF) and translation of the nucleotide sequences was performed using the Open Reading Frame Finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Sequence characteristics of the MsLEA3-1 product were assessed following multiple alignment of deduced amino acid sequences of MsLEA3-1 and the LEA genes from soybean, Glycine max (L.) Merr. (GenBank accession No. AAA91965, CAA80491 and ACU23559, respectively) and castor bean, Ricinus communis L. (GenBank accession No. XP_002519329) using CLUSTALW [15]. The deduced amino acid sequence was analyzed for a potential signal peptide cleavage site using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/), and pI and MW were predicted on the ExPaSy website using Protparam (http://us.expasy.org/tools/pitool.htm).
Sub-cellular localization of MsLEA3-1
The MsLEA3-1 ORF was cloned into the XhoI and SpeI sites of vector pA7-GFP vector, which contains a modified red-shifted green fluorescent protein (GFP) between XbaI and NcoI sites. To amplify the coding sequence of MsLEA3-1, two primers were designed to amplify the fusion protein, one with XhoI restriction site (GFP1) and the other with a SpeI restriction site (GFP2) (Table 1). The empty vector control pA7-GFP and MsLEA-GFP fusion construct was transformed into onion epidermal cells by particle bombardment as described [16]. Cells were incubated for 1 day at 25°C and then observed for transient expression of the MsLEA-GFP fusion protein using a confocal laser scanning microscope.
Real-time RT-PCR for MsLEA3-1
To test the expression of MsLEA3-1, real-time quantitative Q-RT-PCR was performed to amplify MsLEA3-1 on an Opticon II system (Bio-Rad Laboratories/MJ Research, Waltham, MA, USA) using primers RTf and RTr (Table 1 and SuperScript III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. An actin ‘housekeeping’ gene was used to normalize target gene quantities was amplified using primers Actinf and Actinr (Table 1). Salt treatment was carried out on 10-day-old seedlings growing on Hogland’s solution by supplementing with NaCl (150 mM). Total seedling tissue was harvested after 0, 1, 2, 3, 6, 12 and 24 h treatment. ABA treatment was carried out by supplementing the Hogland’s solution with ABA (0.1 mM) at the same stage of development and then harvesting leaves after 0, 1, 3, 6, 8, 16, 24, and 48 h of treatment. Roots, stems and leaves were harvested from 20-day-old seedlings grown under normal greenhouse potting conditions for testing expression of MsLEA3-1 in more mature tissues Total RNA was treated with RNase-free DNase I (Promega) to remove traces of genomic DNA, then the total RNA from each tissue was reverse transcribed as above. The Q-RT-PCR mixtures contained 8 μl of diluted B.carinata cDNA (or 8 μl for control reactions), 10 μl of 2× SYBR Green qPCR Master Mix (catalog no. 11735-040, Invitrogen), and 200 nM of each gene-specific primer in a final volume of 20 μl and reactions were conducted under the following conditions: 2 min at 50°C, followed by 2 min at 95°C, and 40 cycles of 95°C (15 s) and 62°C (30 s). For each pair of primers, gel electrophoresis and melting curve analyses were performed to ensure that only a single PCR amplicon of the expected length and melting temperature was generated. The level of each mRNA was calculated using the mean threshold cycle (Ct) value and normalized to that of the actin reference gene. All results were shown as means of at least three independent RNA extractions (including three technical replicates) with corresponding standard deviations (SD).
Expression of MsLEA3-1 in tobacco plants
To express the alfalfa MsLEA3-1 in tobacco (Nicotiana benthamiana), the MsLEA3-1 cDNA coding region was cloned into pBI121 (Clotech, Japan) by replacing the glucuronidase (GUS) gene, creating plasmid pBI-LE, in which the cauliflower mosaic virus (CaMV) 35S promoter drives the expression of MsLEA3-1 cDNA. Plasmid pBI-LEA was introduced into Agrobacterium tumefaciens LBA4404 by CaCl2 and freezing transformation [17], then introduced into tobacco by co-cultivation [18]. Tobacco cultures were regenerated into plantlets in plastic jars, and then transferred into pots in soil when roots had formed.
The transferred gene was detected in regenerated transgenic tobacco plants by polymerase chain reaction (PCR). Total plant DNA was isolated via the CTAB method from transgenic and non-transgenic plants [19]. To amplify the full coding sequence of MsLEA3-1, two primers were designed, ‘F-coding’ with a BamHI restriction site and ‘R-coding’ with a EcoRI restriction site to amplify a 1,320-bp fragment. PCR was conducted using 30 cycles of 40 s at 94°C (denaturation), 40 s at 60°C (annealing), and 1 min at 72°C (extension) and products were analyzed by electrophoresis on a 1% agarose gel. For real-time quantitative RT-PCR, total RNA was extracted from transgenic and non-transgenic plants using TRIZOL reagent (Invitrogen, USA), and was reverse-transcribed with oligo(dT) primers using M-MLV Reverse Transcriptase (Takara, Japan) in a 20 μl volume. Primers, equipment, conditions, and analysis methods used for Q-RT-PCR analysis of MsLEA3-1 in transgenic tobacco were identical to those used for Q-RT-PCR analysis in alfalfa.
Analysis of salt tolerance of transgenic tobacco plants
T1 selfed seeds from transgenic tobacco plants were germinated in the dark at 25°C on kanamycin-containing (200 mg l−1) MS medium. The surviving seedlings (21-day-old) were transferred to earthenware pots and grown in soil in a controlled environment chamber for an additional period of 3 weeks before they were exposed to stress treatment. At this stage (starting at seven weeks), the green, healthy plants were watered bi-weekly with 240 mM NaCl for a period of 2 weeks and tissue harvested or plants tested at the end of week eight. A 2nd parallel set of plants was also carried through the experiment protocol and similarly watered, but without NaCl (i.e., 0 mM), to serve as an experimental control. For statistical analysis, the mean value of three tested plants within each transgenic line and between each treatment was calculated and means compared with the WT plants using least squares differences by analysis of variance (ANOVA).
Measurement of relative water content (RWC)
Determination of leaf tissue water status was evaluated by calculating the relative water content in fresh leaves of T1 MsLEA3-1 + transgenic tobacco lines and WT plants (three plants each) following the method of Turner [20]. Data was analyzed by ANOVA.
Measurement of electrolyte conductivity
Leaves of T1 transgenic tobacco plants were rinsed with distilled, deionized water to reduce the possibility of ion contamination on their surfaces. Six freshly harvested leaf discs (5-mm diam., two discs per plant) were placed in 20 × 150 mm glass test tubes containing 10 ml of distilled deionized water. The tubes were then shaken at 300 rpm for 4 h in a slanted position under room temperature. Afterward, the initial electrolyte leakage into the solution was monitored with a conductivity meter (DDS SJ-308A, China). Then the solutions were boiled for 20 min to completely lyse plant cell walls and total electrolyte conductivities of the boiled solutions were recorded as absolute conductivity. Percent electrolyte leakage (EL) was calculated by dividing initial conductivity by absolute conductivity and analyzed by ANOVA as above.
Measurement of malondialdehyde (MDA) content
Malondialdehyde (MDA) was estimated in 250 mg fresh T1 transgenic tobacco leaf samples following the procedure of Heath and Packer [21]. Samples were ground in 5 ml distilled water and 5 ml TBA-TCA reagent (0.5% thiobarbituric acid and 20% TCA dissolved in 100 ml of distilled water) was added. The slurry was kept in a water bath at 95°C for 30 min, then cooled by placing in ice bath, and centrifuged at 10,000×g for 10 min. Optical density of the clear supernatant was recorded at 600 and 535 nm. The difference in optical densities gave an actual intensity of color developed as a function of malondialdehyde content. The concentration of MDA was calculated from its extinction coefficient of 155 mM−1 cm−1 and analyzed by ANOVA as above.
Measurement of free proline content
Fresh T1 transgenic tobacco leaf tissue (0.5 g) harvested from each treatment was homogenized in 10 ml of 3% sulphosalicylic acid and filtered through a Whatman No. 2 filter paper. Proline was estimated spectrophotometrically following the ninhydrin method described by Bates [22] using pure proline (Merck) as a standard. Each experiment was repeated at least three times and analyzed by ANOVA as above.
Results
Isolation and sequence analysis of the full-length cDNA of MsLEA3-1
Based on a 409 bp sequence from an alfalfa salt-induced SSH cDNA library, a 1,317 bp fragment from 3′ RACE, and a 434 bp fragment from 5′ RACE, the 1,728 bp full-length cDNA of Medicago sativa MsLEA3-1 protein was amplified and deposited in GenBank (accession no. EU665182). Sequence analysis indicated that MsLEA3-1 cDNA contained an open reading frame of 1,314 bp, a 15 bp ‘-terminal untranslated region (UTR), 263-bp in the 3′-UTR, and had a poly(A) tail (as an on-line supplementary figure A). The ORF encoded a predicted polypeptide of 436 amino acids, had a calculated molecular weight of 47.0 kDa and an estimated pl of 5.18, and a putative signal peptide of 20 amino acids in the N-terminal region. Alignment of deduced amino acid sequences of MsLEA3-1 with LEA proteins from soybean and castor bean using ClustalW revealed that the Medicago sativa L. protein was most similar to the soybean LEA CAA80491, but had showed only 50.87% amino acid sequence identity (as an on-line supplementary figure B).
Subcellular localization of MsLEA3-1
To examine the subcellular localization of MsLEA3-1, a fusion protein was constructed between MsLEA3-1 and a GFP, The recombinant plasmid and the pA7-GFP vector were transformed into onion epidermal cells by a gene gun. When the cultured onion epidermal cells with the MsLEA-GFP fusion were examined after 1 day at room temperature by confocal microscopy, a strong fluorescence signal was observed only surrounding the nucleolus (Fig. 1d, e, f). In contrast, the GFP signal was distributed throughout the onion epidermal cells in tissues transformed with the control pA7-GFP vector (Fig. 1a, b, c). Together with protein sequence analysis, these results suggest that MsLEA3-1 is a nucleolar-membrane localized protein.
Subcellular localization of MsLEA3-1 in onion epidermal cells. The MsLEA-GFP fusion and the pA7-GFP control plasmid were introduced into the onion cells using a gene gun and fluorescence signals were examined by a confocal laser scanning microscope. a–c GFP fluorescence distributed throughout the entire cells from the GFP empty vector. d–f GFP fluorescence from cells expressing MsLEA-GFP fusion protein localized to the nucleolus. a, d Dark field vision. b, e Bright light vision. c, f Superposition of bright and dark visions
MsLEA3-1 expression pattern analysis
In order to determine the expression pattern of MsLEA3-1, real-time Q-PCR was carried out to examine the transcript levels of MsLEA3-1 in Medicago sativa. Organ-specific expression of MsLEA3-1 occurred in more mature plants, such that we found that transcripts of MsLEA3-1 were 4.5-fold more abundant in leaves than in stems and not detectable in roots (Fig. 2a). Under NaCl treatment, the gene expressed differentially in seedling leaves. Here, upregulation was detected as early as 1 h after treatment with salt, and the mRNA level was substantially elevated at 12 h. Q-PCR analysis recorded two expression maxima for this gene, one at 1 h and the other at 12 h, before expression declined at 24 (Fig. 2b). In response to ABA treatment, the expression pattern of MsLEA3-1 was quite distinct from response to salt stress. MsLEA3-1 transcripts increased only slightly by 3 h, in seedling leaves with ABA treatment, but then increased rapidly and substantially by 6 h and reached a maximum titre in the leaves by the end of the testing period (48 h) (Fig. 2c). These results strongly suggested that MsLEA3-1 is responsive to environmental stress and hormone and should be tested to see whether it can regulate plant responses to stress.
Expression patterns of MsLEA3-1 in three organs and in response to NaCl and ABA treatments. a Roots, stem and leaves of 20-day-old plants under non-stress conditions. b Seeding leaves (10-days-old) under 24 h treatment with 150 mM NaCl. c Seeding leaves (10-days-old) under 48 h treatment with 0.1 mM ABA
Expression of MsLEA3-1 confers tolerance to salt stress in transgenic tobacco
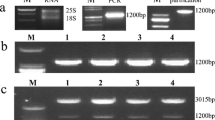
In order to investigate the role of MsLEA3-1 in response to salt stress in planta, we generated transgenic tobacco plants with MsLEA3-1 under the control of the CaMV 35S promoter. Independently generated putative transformants (24) were verified by PCR using MsLEA3-1-specific primers (Table 1). Nine of these plants were confirmed to have the expected 1,314 bp amplified product, whereas non-transgenic control plants showed no bands (Fig. 3a).
Confirmation and expression of transformants from transgenic tobacco plants using MsLEA3-1 specific primers (Table 1). a Confirmation of transgenic plants using RT-PCR. NT non-transgenic plant. Lanes 1 to 9, transgenic plants. M DL2000plus size marker. b Expression of MsLEA3-1 in six T1 transgenic tobacco plants by Q-PCR analysis. Expression is relative to non-transgenic
Expression of MsLEA3-1 in the leaves of six T1 tobacco transformants was analyzed by real-time quantitative RT-PCR. All six tested transgenic plants were confirmed to express the MsLEA3-1 gene, while the non-transgenics did not (Fig. 3b). Moreover, the level of expression varied by line, with plants from Lines 2 and 4 showing the highest levels of transcript compared with only weak expression in Lines 1 and 5. The other two lines (3 and 6) had intermediate levels of expression.
To investigate whether constitutive expression of the MsLEA3-1 gene in tobacco might provide protection against salt stress, relative water content (RWC), electrical leakage (EL), malonyldialdehyde (MDA) content and proline content were measured in eight transgenic lines and one non-transgenic line before and after 240 mM NaCl. Under untreated conditions, no differences were observed for these four parameters among any of the genotypes. However, MDA content did increase in all plant lines after exposure to salt. This rise in MDA was much less for several of the transgenic plants compared with the 3-fold increase in MDA in the non-transgenic plants after salt exposure (Fig. 4a).
Salt stress tolerance in MsLEA3-1 expressing transgenic tobacco plants. a Relative water content. b Electrolyte leakage. c MDA content. d Proline content. NT non-transgenic control plants. Lines 1 to 8, Transgenic tobacco plants. All plants were treated with 240 mM NaCl. Each value represents the mean of measurements from three individual plants; error bars indicate standard error of the means (SE). Letters indicate significant differences of the means (P < 0.05)
For example, MDA content was lowest (58.3%) (P < 0.05) compared with the non-transgenic plants after NaCl treatment. These results infer that the heterologous MsLEA3-1 gene product can function to protect the transgenic tobacco plants against oxidative stress.
After treatment with 240 mM NaCl, relative water content decreased in all genotypes, but especially in the non-transgenic plants (Fig. 4b). In contrast, the RWC in transgenic Lines 2 and 8 were 10.0 and 8.5% higher and the most responsive lines 1, 5 and 6 were 23.7, 22.5 and 21.5% higher (P < 0.05), respectively, than the non-transgenic line (Fig. 4b). These results suggest that the MsLEA3-1 transgene product possibly prevents water loss during salinity stress and may protect against the crystallization of cellular components.
Although electrolyte leakage was increased in all plants after treatment, leakage from non-transgenics (46.5%) was highest of all plants tested. EL from transgenic Line 5 (23.2%), Line 7 (29.4%), Line 2 (40.3%), and Line 4 (35.5%) was substantially lower than leakage from the non-transgenic plants (Fig. 4c). Analysis of variance indicated significant differences even between the transgenics, and the EL for Line 5 was the least, being 50.2% (P < 0.05) lower than non-transgenic plants. The implication of these experiments is that the plasma membrane from plants of line 5 likely suffered only slight damage. This suggests that the MsLEA3-1 gene product may function to generate protection for cell membranes, while serving to enhance the salt tolerance of transgenic plants.
Under salt stress, proline accumulation in all transgenic lines was substantially higher than in the non-transgenic line (Fig. 4d). In Lines 1, 2 and 7, the leaf proline content was more than 4-fold higher than that recorded in control plants, while more than 5-fold higher in Lines 3, 4, 5, 6 and 8. These data suggest that the MsLEA3-1 gene product may function to induce proline, a compatible osmolyte, as a part of their mechanism at increasing the salt tolerance of transgenic plants.
Discussion
Alfalfa (Medicago sativa L.), the most important perennial forage crop in the world, has a wide distribution in irrigated arid and semi-arid regions all over the world. With the development of genetic engineering, it has become feasible to enhance the tolerance to abiotic stress in crops using these techniques. There are numerous studies where heterologous LEA protein genes have been introduced into plants or microorganisms and then showed a degree of increased stress tolerance. LEA genes have been cloned from many plant species [23–25], but their cloning and molecular characterization have not previously been reported in alfalfa. With this current study, we have now become the first group to isolate a LEA gene from Medicago sativa L. Our analysis showed that the MsLEA3-1 protein shares only a low level of amino acid sequence similarity to other LEA proteins in the NCBI database and is closest in homology to LEA from soybean, indicating that it is a novel LEA gene.
Abiotic stresses such as drought, high salinity, and freezing, disturb the water balance of cells, affect plant growth, and thereby affect crop productivity [26]. So far, researches on LEA proteins have mainly focused on salt stress response pathway in both eukaryotic and procaryotic organisms. However, it is generally believed that a number of molecular genetic events are co-ordinated through the action of the plant growth regulator, abscisic acid (ABA) [27]. Therefore, we examined changes in MsLEA3-1 leaf expression after the alfalfa plants were subjected to treatment with NaCl or ABA. We demonstrated that MsLEA3-1 was transiently induced in two peaks of expression by NaCl before declining at 24 h. In contrast, MsLEA3-1 transcripts accumulated slowly and continuously under ABA stress, reaching a maximal level at the end of the 48 h test period in the salinity expression pattern is distinct from the expression pattern of CaLEA6 [28]. It implies that the MsLEA3-1 may be involved in the response of alfalfa to salt stress. The identification and correlation of expression of additional genes in these MsLEA3-1 transcriptional responses to salt and ABA should give us additional information to determine whether MsLEA3-1 acts directly or indirectly to regulate the salt and ABA stress response.
LEA proteins are members of a large group of hydrophilic, glycine-rich proteins found in plants [29]. The exact functions of LEA proteins are not known. However, they are thought to function through the maintenance of protein or membrane structure, sequestration of ions, the binding of water, or as molecular chaperones to help prevent the formation of damaging protein aggregates [30–32]. In this study, we found that the Medicago sativa L. LEA gene enhanced salt tolerance when expressed in heterologous transgenic tobacco, confirming that this MsLEA3-1 gene is a stress-tolerance gene and can function in more than just its own species. The salt stress tests show that MD and electrical leakage are all significantly lower in the transgenic plants than in non-transgenic plants. This indicates that the introduction of the LEA gene protected cell membranes and decreased membrane damage under saline conditions and is consistent with the findings of Wang et al. [33]. The relative water content and proline content also accumulated much more in transgenic tobacco compared with the non-transgenic plants. These results suggest that the mechanism of salt tolerance conferred by the LEA gene product may be through osmotic adjustment and protection of membrane stability, which is similar to Maggio [34].
In conclusion, a gene coding for a salt-induced protein MsLEA3-1 was isolated from Medicago sativa. The protein is localized in the nucleolus, had a signal peptide at the N-terminal end, and could be induced by salt and ABA. Development of transgenic tobacco with the cloned MsLEA3-1 gene demonstrated that it could enhance salt tolerance in heterologous transgenic plants. The discovery of this new member of the LEA gene family will contribute to the study of mechanisms of salt tolerance in Medicago sativa L., and will also provide an additional gene in the toolbox for the genetic improvement of stress tolerance in crop plants.
References
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–420
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2:503–512
Vicient CM, Hull G, Guilleminot J, Devic M, Delseny M (2000) Differential expression of the Arabidopsis genes coding for Em-like proteins. J Exp Bot 51:1211–1220
Dure L III (1993) A repeating 11-mer amino acid motif and plant desiccation. Plant J 3:363–369
Ramanjulu S, Bartels D (2002) Drought and desiccation-induced modulation of gene expression in plants. Plant Cell Environ 25:141–151
Imai R, Chang L, Ohta A, Bray EA, Takagi M (1996) A lea-class gene of tomato confers salt and freezing tolerance when expressed in Saccharomyces cerevisiae. Gene 170:243–248
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice. Oryza sativa L., encode transcription activators that function in drought, high salt and cold responsive gene expression. Plant J 33:751–763
Honjoh KI, Oda Y, Takata R, Miyamoto T, Hatano S (1999) Introduction of the hiC6 gene, which encodes a homologue of a late embryogenesis abundant (LEA) protein, enhances freezing tolerance of yeast. J Plant Physiol 155:509–512
Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110:249–257
Rohila JS, Jain RK, Wu R (2002) Genetic improvement of Basmati rice for salt and drought tolerance by regulated expression of a barley Hva1 cDNA. Plant Sci 163:525–532
Babu RC, Zhang J, Blum A, Ho DTH, Wu R, Nguyen HT (2004) HVA1. a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166:855–862
Jin HC, Sun Y, Yang QC, Chao YH, Kang JM, Jin H, Li Y, Margaret G (2010) Screening of genes induced by salt stress from Alfalfa. Mol Biol Rep 37:745
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
CSH Protocols; 2007;10.1101/pdb.prot4689
Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Horsch RB, Fry J, Hoffmann N, Niedermeyer J, Rogers SG, Fraley TF (1988) Leaf disc transformation. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer, Dordrecht, pp 1–9
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Turner NC (1981) Techniques and experimental approaches for the measurement of plant water status. Plant Soil 58:339–366
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Raynal M, Gaubier P, Grellet F, Delseny M (1990) Nucleotide sequence of a radish cDNA clone coding for a late embryogenesis abundant (LEA) protein. Nucl Acids Res 18:6132
Moons A, de KA, van MM (1997) A group 3 LEA cDNA of rice, responsive to abscisic acid, but not to jasmonic acid, shows variety-specific differences in salt stress response. Gene 191:197–204
Shih MD, Lin SC, Hsieh JS, Tsou CH, Chow TY, Lin TP, Hsing YI (2004) Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol Biol 56:689–703
Yamaguchi-Shinozaki K, Kasuga M, Liu Q, Nakashima K, Sakuma Y, Abe H, Shinwari ZK, Seki M, Shinozaki K (2002) Biological mechanisms of drought stress response. JIRCAS Work Rep 23:1–8
Ramanjulu S, Bartels D (2002) Drought and desiccation induced modulation of gene expression in plants. Plant Cell Environ 25:141–151
Kim HS, Lee JH, Kim JJ, Kim CH, Jun SS, Hong YN (2005) Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum. Gene 344:115–123
Soulages JL, Kim K, Arres EL, Walters C, Cushman JC (2003) Conformation of a group 2 late embryogenesis abundant protein from soybean. Evidence of poly(l-proline)-type II structure. Plant Physiol 131:963–975
Close TJ (1997) Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant 100:291–296
Browne J, Tunnacliffe A, Burnell A (2002) Plant desiccation gene found in a nematode. Nature 416:38
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157
Wang YC, Jiang J, Zhao X, Liu GF, Yang CP, Zhan LP (2006) A novel LEA gene from Tamarix and rossowii confers drought tolerance in transgenic tobacco. Plant Sci 171:655–662
Maggio A, Reddy MP, Joly RJ (2000) Leaf gas exchange and solute accumulation in the halophyte Salvadora persica grown at moderate salt. Environ Exp Bot 44:31–38
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bai, Y., Yang, Q., Kang, J. et al. Isolation and functional characterization of a Medicago sativa L. gene, MsLEA3-1 . Mol Biol Rep 39, 2883–2892 (2012). https://doi.org/10.1007/s11033-011-1048-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1048-z