Abstract
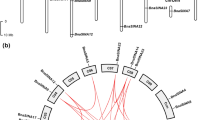
Six full-length cDNA encoding boron transporters (BOR) were isolated from Brassica napus (AACC) by rapid amplification of cDNA ends (RACE). The phylogenic analysis revealed that the six BORs were the orthologues of AtBOR1, which formed companying with the triplication and allotetra-ploidization process of B. napus, and were divided into three groups in B. napus. Each group was comprised of two members, one of which was originated from Brassica rapa (AA) and the other from Brassica oleracea (CC). Based on the phylogenetic relationships, the six genes were named as BnBOR1;1a, BnBOR1;1c, BnBOR1;2a, BnBOR1;2c, BnBOR1;3a and BnBOR1;3c, respectively. The deduced BnBOR1 s had extensive similarity with other plant BORs, with the identity of 74–96.8% in amino acid sequence. The BnBOR1;3a and BnBOR1;3c resembled AtBOR1 in number and positions of the 11 introns, but the others only have 9 introns. After the gene duplication, there was evidence of purifying selection under a divergent selective pressure. The expression patterns of the six BnBOR1 s were detected by semi-quantitative RT–PCR. The BnBOR1;3a and BnBOR1;3c showed a ubiquitous expression in all of the investigated tissues, whereas the other four genes showed similar tissue-specific expression profile. Unlike the non-transcriptional regulation of AtBOR1, the expression of BnBOR1;1c and BnBOR1;2a were obviously induced by boron deficiency. This study suggested that the BOR1 s had undergone a divergent expression pattern in the genome of B. napus after that the B. napus diverged from Arabidopsis thaliana.



Similar content being viewed by others
Abbreviations
- B:
-
Boron
- MIPs:
-
Major intrinsic proteins
- RACE:
-
Rapid amplification of cDNA ends
- MYA:
-
Million years ago
- CDS:
-
Coding sequence
- PCR:
-
Polymerase chain reactions
- RT–PCR:
-
Reverse transcription polymerase chain reactions
- LRT:
-
Likelihood ratio test
- TR:
-
Two ration model
- HWSB:
-
Hot water-soluble boron
References
Warington K (1923) The effect of boric acid and borax on the broad bean and certain other plants. Ann Bot 37(4):629–672
Nielsen FH (2000) The emergence of boron as nutritionally important throughout the life cycle. Nutrition 16(7–8):512–514. doi:10.1016/S0899-9007(00)00324-5
Kobayashi M, Matoh T, Azuma J-i (1996) Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol 110(3):1017–1020
Ishii T, Matsunaga T (1996) Isolation and characterization of a boron-rhamnogalacturonan-II complex from cell walls of sugar beet pulp. Carbohydr Res 284(1):1–9. doi:10.1016/008-6215(96)00010-9
O’Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294(5543):846–849. doi:10.1126/science.1062319
O’Neill MA, Ishii T, Albersheim P, Darvill AG (2004) Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol 55:109–139. doi:10.1146/annurev.arplant.55.031903.141750
Marschner H (1995) Functions of mineral nutrients: micronutrients. In: Mineral nutrition of higher plants, 2nd ed. Academic Press, London, pp 313–404
Hu H, Brown PH, Labavitch JM (1996) Species variability in boron requirement is correlated with cell wall pectin. J Exp Bot 47(2):227–232. doi:10.1093/jxb/47.2.227
Raven JA (1980) Short- and long-distance transport of boric acid in plants. New Phytol 84:231–249
Brown PH, Shelp BJ (1997) Boron mobility in plants. Plant Soil 193(1):85–101
Dordas C, Brown PH (2000) Permeability of boric acid across lipid bilayers and factors affecting it. J Membrane Biol 175(2):95–105. doi:10.1007/s002320001058
Dannel F, Pfeffer H, Römheld V (2000) Characterization of root boron pools, boron uptake and boron translocation in sunflower using the stable isotopes 10B and 11B. Aust J Plant Physiol 27(5):397–405. doi:10.1071/PP99086
Stangoulis JC, Reid RJ, Brown PH, Graham RD (2001) Kinetic analysis of boron transport in Chara. Planta 213(1):142–146. doi:10.1007/s004250000484
Dordas C, Brown PH (2001) Evidence for channel mediated transport of boric acid in squash (Cucurbita pepo). Plant Soil 235(1):95–103. doi:10.1023/A:1011837903688
Dordas C, Chrispeels MJ, Brown PH (2000) Permeability and channel-mediated transport of boric acid across membrane vesicles isolated from squash roots. Plant Physiol 124(3):1349–1362
Nozawa A, Takano J, Kobayashi M, Wirén NV, Fujiwara T (2006) Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett 262(2):216–222. doi:10.1111/j.1574-6968.2006.00395.x
Takano J, Wada M, Ludewig U, Schaaf G, Wirén NV, Fujiwara T (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18(6):1498–1509. doi:10.1105/tpc.106.041640
Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T (2008) NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20(10):2860–2875. doi:10.1105/tpc.108.058628
Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420(6913):337–340. doi:10.1038/nature01139
Tanaka M, Fujiwara T (2008) Physiological roles and transport mechanisms of boron: perspectives from plants. Pflügers Archiv 456(4):671–677. doi:10.1007/s00424-007-0370-8
Takano J, Miwa K, Yuan L, Wirén NV, Fujiwara T (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102(34):12276–12281. doi:10.1073/pnas.0502060102
Nakagawa Y, Hanaoka H, Kobayashi M, Miyoshi K, Miwa K, Fujiwara T (2007) Cell-type specificity of the expression of Os BOR1, a rice efflux boron transporter gene, is regulated in response to boron availability for efficient boron uptake and xylem loading. Plant Cell 19(8):2624–2635. doi:10.1105/tpc.106.049015
Sutton T, Baumann U, Hayes J, Collins NC, Shi B-J, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, Langridge P (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318(5855):1446–1449. doi:10.1126/science.1146853
Miwa K, Takano J, Omori H, Seki M, Shinozaki K, Fujiwara T (2007) Plants tolerant of high boron levels. Science 318(5855):1417. doi:10.1126/science.1146634
Takano J, Toyoda A, Kasai K, Miwa K, Fuji K, Fujiwara T (2008) Endocytic degradation and polarized localization of borate transporters dependent on sorting motifs. In: 19th International Conference on Arabidopsis Research. Montreal, pp 197–198
Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA 107(11):5220–5225. doi:10.1073/pnas.0910744107
Wolf BF, Wirén NV (2002) Plant biology: ping-pong with boron. Nature 420(6913):282–283. doi:10.1038/420282a
Park M, Li Q, Shcheynikov N, Zeng W, Muallem S (2004) NaBC1 is a ubiquitous electrogenic Na+-coupled borate transporter essential for cellular boron homeostasis and cell growth and proliferation. Mol Cell 16(3):331–341. doi:10.1016/j.molcel.2004.09.030
Kaya A, Karakaya HC, Fomenko DE, Gladyshev VN, Koc A (2009) Identification of a novel system for boron transport: Atr1 is a main boron exporter in yeast. Mol Cell Biol 29(13):3665–3674. doi:10.1128/MCB.01646-08
Franzke A, German D, Al-Shehbaz IA, Mummenhoff K (2009) Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58(2):425–437
U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:385–452
Parkin IA, Sharpe AG, Lydiate DJ (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46(2):291–303. doi:10.1139/g03-006g03-006
Wuding L (1995) Microelement nutrition and fertilization in china. China Agriculture Press, Beijing, pp 8–36
Yuai Y, Jianming X, Zhengqiang Y, Ke W (1993) Responses of rape genotypes to boron application. Plant Soil 155–156(1):321–324. doi:10.1007/BF00025047
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81(24):8014–8018
Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y (1998) Predicting function: from genes to genomes and back. J Mol Biol 283(4):707–725. doi:10.1006/jmbi.1998.2144
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–98
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599. doi:10.1093/molbev/msm092
Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18(3):502–504
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591. doi:10.1093/molbev/msm088
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11(5):725–736
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15(5):568–573
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155(1):431–449
Nielsen R, Yang Z (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148(3):929–936
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157(1):105–132. doi:10.1016/0022-2836(82)90515-0
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11(6):681–684
Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14(6):1347–1357
Schaaf G, Schikora A, Haberle J, Vert G, Ludewig U, Briat JF, Curie C, von Wiren N (2005) A putative function for the arabidopsis Fe-Phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol 46(5):762–774. doi:10.1093/pcp/pci081
Li WH, Gojobori T (1983) Rapid evolution of goat and sheep globin genes following gene duplication. Mol Biol Evol 1(1):94–108
Nei M (2005) Selectionism and neutralism in molecular evolution. Mol Biol Evol 22(12):2318–2342. doi:10.1093/molbev/msi242
Yang Z (2002) Inference of selection from multiple species alignments. Curr Opin Genet Dev 12(6):688–694. doi:10.1016/S0959-437X(02)00348-9
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30771283, 30971861) and the National 863 High Technology Program of China (2007AA10Z117).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2011_930_MOESM1_ESM.png
Supplementary figure Fig. S1 Multiple alignment of BnBOR1 s with AtBOR1 (NP_850469) Dots indicate identical residues with the BnBOR1;1a; grey box and star indicate possible positive selective sites detected by M8 model in PAml analysis; line indicates the conserved HCO3_cotransporer domain (PNG 160 kb)
Rights and permissions
About this article
Cite this article
Sun, J., Shi, L., Zhang, C. et al. Cloning and characterization of boron transporters in Brassica napus . Mol Biol Rep 39, 1963–1973 (2012). https://doi.org/10.1007/s11033-011-0930-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0930-z