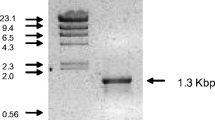
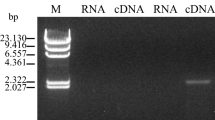
Abstract
The 972 bp length of transaldolase gene tal was cloned from Pichia stipitis CICC1960, encoding a 323 amino acid protein with a calculated molecular mass of 35.36 kDa and isoelectric point of 5.20. Real time PCR analysis demonstrated that the mRNA transcript level of constitutive tal gene rise on xylose, glucose, fructose, mannose, galactose and sucrose as carbon source, respectively. Furthermore, the transcription of tal gene in P. stipitis on xylose was higher than on other carbon source, indicating that transaldolase plays a part in xylose utilization. To deeply study the tal gene biological function, it was expressed in Fusarium oxysporum CCTCC M209040. Recombinant transaldolase activity of transformant F. oxysporum M209040-Tal2 was about 0.52 U mg−1 protein and was 1.57 times higher than that of the wild type F. oxysporum CCTCC M209040, indicating that the improvement of transaldolase activity in transformant was due to expression of the exogenous tal gene. Growth of transformant F. oxysporum M209040-Tal2 without selection pressure did not affect the level of hygromycin resistance of the transformants, suggesting that integrated tal gene was stable in mitosis. Fermentation trials of F. oxysporum M209040-Tal2 showed that the ethanol yield improved by 8.39 and 11.71% on glucose and xylose substrates, respectively, demonstrating that the expression of tal gene from P. stipitis CICC1960 in F. oxysporum CCTCC M209040 could improve ethanol production.







Similar content being viewed by others
References
Lu JL, Liao JC (1997) Metabolic engineering and control analysis for production of aromatics: role of transaldolase. Biotechnol Bioeng 53:132–138. doi:10.1002/(SICI)1097-0290(19970120)53:2<132:AID-BIT2>3.0.CO;2-P
Jin YS, Alper H, Yang YT, Stephanopoulos G (2005) Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl Environ Microb 71:8249–8256. doi:10.1128/AEM.71.12.8249-8256.2005
Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S (1995) Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240–243. doi:10.1126/science.267.5195.240
Yanase H, Sato D, Yamamoto K et al (2009) Genetic engineering of Zymobacter palmae for production of ethanol from xylose. Appl Environ Microbiol 73:2592–2599. doi:10.1128/AEM.02302-06
Singh A, Kumar PKR (1991) Fusarium oxysporum: status in bioethanol production. Crit Rev Biotechnol 11:129–147. doi:10.3109/07388559109040619
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Nat Technol 9:963–967. doi:10.1038/nbt1091-963
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Chen PY, Wang CK, Song SC, To KY (2003) Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol Breed 11:287–293. doi:10.1023/A:1023475710642
Covert SF, Kapoor P, Lee Mh, Briley A, Nairn CJ (2001) Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res 105:259–264. doi:10.1017/S0953756201003872
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C (2003) The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Technol 14(1):33–43
Li J, Lu YM, Xue LX, Xie H (2010) A structurally novel salt-regulated promoter of duplicated carbonic amhydrase gene 1 from Dunaliella salina. Mol Biol Rep 37:1143–1154. doi:10.1007/s11033-009-9901-z
Wang W, Tai FJ, Chen SN (2008) Optimizing protein extraction from plant tissues for enhanced proteomics analysis. J Sep Sci 31:2032–2039. doi:10.1002/jssc.200800087
Soderberg T, Alver RC (2004) Transaldolase of Methanocaldococcus jannaschii. Archaea 1:255–262
Bradforda MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of dye-binding. Anal Biochem 72:248–254
Zhao L, Zhang X, Tan T (2008) Influence of various glucose/xylose mixtures on ethanol production by Pachysolen tannophilus. Biomass Bioenergy. doi: 10.1016/j.biombioe.2008.02.011
Gefflaut T, Blonski C, Perie J, Willson M (1995) Class I aldolases: substrate specificity, mechanism, inhibitors and structural aspects. Prog Biophys Mol Biol 63:301–340
Fiki AE, Metabteb GE, Bellebna C, Wartmann T et al (2007) The Arxula adeninivorans ATAL gene encoding transaldolase-gene characterization and biotechnological exploitation. Appl Microbiol Biotechnol 74:1292–1299. doi:10.1007/s00253-006-0785-8
Wang T, Merja P, Gao P (1999) Expression of xylose-metabolic key genes of Trichoderma reesei on various carbon sources measured by a series of Northern hybridizations. ACTA Microbiol Sin 39:503–509
Wahlbom CF, Otero RRC, van Zyl WH, Hahn-Hägerdal B, Jönsson LJ (2003) Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Appl Environ Microbiol 69:740–746. doi:10.1128/AEM.69.2.740-746.2003
Vatanaviboon P, Varaluksit T, seeanukun C, Mongkolsuk S (2002) Transaldolase exhibits a protective role against menadione toxicity in Xanthomonas campestris pv. phaseoli. Biochem Biophys Res Commun 297:968–973. doi:10.1016/S0006-291X(02)02329-X
Panagiotou G, Christakopoulos P, Olsson L (2005) Simultaneous saccharification and fermentation of cellulose by Fusarium oxysporum F3 growth characteristics and metabolite profiling. Enzyme Microb Technol 36:693–699. doi:10.1016/j.enzmictec.2004.12.029
Christakopoulos P, Kekos D, Macris BJ (1995) Purification and mode of action of a low molecular mass endo-1, 4-β-d-glucanase from Fusarium oxysporum. J Biotechnol 39:85–93. doi:10.1016/0168-1656(94)00147-5
Christakopoulos P, Mamma D, Nerinckx W (1995) Purification and characterization of a less randomly acting endo-1, 4-β-d-glucanase from the culture fitrates of Fusarium oxysporum. Arch Biochem Biophys 316:428–433
Moukouli M, Topakas E, Christakopoulos P (2008) Cloning, characterization and functional expression of an alkalitolerant type C feruloyl esterase from Fusarium oxysporum. Appl Microbiol Biotechnol 79:245–254. doi:10.1007/s00253-008-1432-3
Panagiotou G, Christakopoulos P, Villas-Boas SG, Olsson L (2005) Fermentation performance and intracellular metabolite profiling of Fusarium oxysporum cultivated on a glucose-xylose mixture. Enzym Microb Technol 36:100–106. doi:10.1016/j.enzmictec.2004.07.009
Panagiotou G, Christakopoulos P (2004) NADPH-dependent d-aldose reductases and xylose fermentation in Fusarium oxysporum. Biosci Bioeng 97:299–304. doi:10.1016/S1389-1723(04)70209-1
Panagiotou G, Kekos D, Macris B, Christakopoulos P (2002) Purification and characterisation of NAD(+)-dependent xylitol dehydrogenase from Fusarium oxysporum. Biotechnol Lett 24:2089–2092. doi:10.1023/A:1021317614948
Kourtoglou E, Mamma D, Topakas E, Christakopoulos P (2008) Purification, characterization and mass spectrometric sequencing of transaldolase from Fusarium oxysporum. Process Biochem 43:1094–1101. doi:10.1016/j.procbio.2008.05.013
Gouka RJ, Gerk C, Hooykaas JJ, Wouter CT, Groot JA (1999) Transformation of Aspergillus awamori by Agrobacterium tumefaciens homologous recombination. Nat Biotechnol 17:598–601. doi:10.1038/9915
Zwiers LH, Waard MAD (2001) Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella grominicola. Curr Genet 39:388–393. doi:10.1007/s002940100216
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17. doi:10.1007/s00294-005-0578-0
Zeng FS, Qian JJ, Kang J, Wang HY et al (2009) Histochemical study of β-glucuronidase activity in transgenic birch. Chin Bull Bot 44:484–490. doi:10.3969/j.issn.1674-3466.2009.04.010
Wang Y C, Qu GZ, Li H Y et al (2010) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol Biol Rep 37:1119–1124. doi:10.1007/s11033-009-9884-9
Feng SY, Xue LX, Liu HT, Lu PJ (2009) Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol Biol Rep 36:1433–1439. doi:10.1007/s11033-008-9333-1
Sun L, Cai H, Xu W, Hu Y, Lin Z (2002) CaMV 35S promoter directs β-glucuronidase expression in Ganoderma lucidum and Pleurotus citrinopileatus. Mol Biotechnol 20:239–244. doi:10.1385/MB:20:3:239
Meinander NQ, Boels I, Hahn-Hagerdal B (1999) Fermentation of xylose/glucose mixtures by metabolically engineered Saccharomyces cerevisiae strains expressing XYL1 and XYL2 from Pichia stipitis with and without overexpression of TAL1. Bioresour Technol 68:79–87
Gu Y, Li J, Zhang L et al (2009) Improvement of xylose utilization in Clostridium acetobutylicum via expression of the talA gene encoding transaldolase from Escherichia coli. J Biotechnol 143:284–287. doi:10.1016/j.jbiotec.2009.08.009
Acknowledgments
This research was funded by National 863 Research Program (2006AA10Z424) and National Key Technology R&D Program of China during the eleventh 5-year plan period (2006BAD07A01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, Jx., Yang, Q., Liu, Zh. et al. The characterization of transaldolase gene tal from Pichia stipitis and its heterologous expression in Fusarium oxysporum . Mol Biol Rep 38, 1831–1840 (2011). https://doi.org/10.1007/s11033-010-0299-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0299-4