Abstract
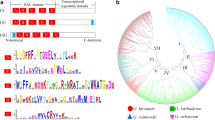
Annexins constitute a family of multifunction and structurally related proteins. These proteins are ubiquitous in the plant kingdom, and are important calcium-dependent membrane-binding proteins that participate in the polar development of different plant regions such as rhizoids, root caps, and pollen tube tips. In this study, a novel cotton annexin gene (designated as GhFAnnx) was isolated from a fiber cDNA library of cotton (Gossypium hirsutum). The full-length cDNA of GhFAnnx comprises an open reading frame of 945 bp that encodes a 314-amino acid protein with a calculated molecular mass of 35.7 kDa and an isoelectric point of 6.49. Genomic GhFAnnx sequences from different cotton species, TM-1, Hai7124 and two diploid progenitor cottons, G. herbaceum (A-genome) and G. raimondii (D-genome) showed that at least two copies of the GhFAnnx gene, each with six exons and five introns in the coding region, were identified in the allotetraploid cotton genome. The GhFAnnx gene cloned from the cDNA library in this study was mapped to the chromosome 10 of the A-subgenome of the tetraploid cotton. Sequence alignment revealed that GhFAnnx contained four repeats of 70 amino acids. Semi-quantitative reverse transcriptase-polymerase chain reaction revealed that GhFAnnx is preferentially expressed in different developmental fibers but its expression is low in roots, stems, and leaves. Subcellular localization of GhFAnnx in onion epidermal cells and cotton fibers suggests that this protein is ubiquitous in the epidermal cells of onion, but assembles at the edge and the inner side of the apex of the cotton fiber tips with brilliant spots. In summary, GhFAnnx influences fiber development and is associated with the polar expansion of the cotton fiber during elongation stages.




Similar content being viewed by others
References
Delmer DP, Potikha TS (1997) Structures and functions of annexins in plants. Cell Mol Life Sci 53:546–553
Creutz CE, Pazoles CJ, Pollard HB (1978) Identification and purification of an adrenal medullary protein (synexin) that causes calcium-dependent aggregation of isolated chromaffin granules. J Biol Chem 253:2858–2866
Boustead CM, Smallwood M, Small H, Bowles DJ, Walker JH (1989) Identification of Ca2+-dependent phospholipid-binding proteins in higher plant cells. FEBS Lett 244:456–460
Konopka-Postupolska D (2007) Annexins: putative linkers in dynamic membrane–cytoskeleton interactions in plant cells. Protoplasma 230:203–215
Andrawis A, Solomon M, Delmer DP (1993) Cotton fiber annexins: a potential role in the regulation of callose synthase. Plant J 3:763–772
Potikha TS, Delmer DP (1997) cDNA clones for annexin AnnGh1 (accession no. U73746) and AnnGh2 (accession no. U73747) from Gossypium hirsutum (cotton)1 (PGR 97-003). Plant Physiol 113:305
Shin H, Brown RM (1999) GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiol 119:925–934
Hofmann A, Delmer DP, Wlodawer A (2003) The crystal structure of annexin Gh1 from Gossypium hirsutum reveals an unusual S3 cluster-implications for cellulose synthase complex formation and oxidative stress response. Eur J Biochem 270:2557–2564
Blackbourn HD, Barker PJ, Huskisson NS, Battey NH (1992) Properties and partial protein sequence of plant annexins. Plant Physiol 99:864–871
Blackbourn HD, Battey NH (1993) Annexin-mediated secretory vesicle aggregation in plants. Plant Physiol 89:27–32
Carroll AD, Moyen C, Van Kesteren P, Tooke F, Battey NH, Brownlee C (1998) Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. Plant Cell 10:1267–1276
Bassani M, Neumann PM, Gepstein S (2004) Differential expression profiles of growth-related genes in the elongation zone of maize primary roots. Plant Mol Biol 56:367–380
Clark GB, Dauwalder M, Roux SJ (1992) Purification and immunolocalization of annexin-like protein in pea seedlings. Planta 187:1–9
Clark GB, Dauwalder M, Roux SJ (1994) Immunolocalization of an annexin-like protein in corn. Adv Space Res 14:341–346
Clark GB, Turnwald S, Tirlapur UK, von der Mark K, Roux SJ, Scheuerlein R (1995) Induction and polar distribution of annexin-like proteins during phytochrome-mediated rhizoid initiation and growth in spores of the ferns Dryopteris and Anemia. Planta 197:376–384
Clark GB, Lee DW, Dauwalder M, Roux SJ (2005) Immunolocalization and histochemical evidence for the association of two different Arabidopsis annexins with secretion during early seedling growth and development. Planta 220:621–631
Jiang JX, Zhang TZ (2003) Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method. Cotton Sci 15:166–167
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Nicholas KB, Nicholas HB Jr (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author. Available at http://www.psc.edu/biomed/genedoc
Han ZG, Guo WZ, Song XL (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Genomics 272:308–327
Guo WZ, Cai CP, Wang CB, Zhao L, Wang L, Zhang TZ (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics 9:314
Van-Ooijen JW, Voorrips RE (2001) JoinMap version 3.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen
Liu RH, Meng JL (2003) MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25:317–321
Burgoyne RD, Geisow MJ (1989) The annexin family of calcium-binding proteins. Cell Calcium 10:1–10
Wang GL, Yang HY, Xia R, Fang HJ, Jing SX (2001) Cloning and sequencing the full-length cDNA of annexin from strawberry fruit. Acta Bot Sin 43:874–876
Anuphon L, Jennifer CM, Vadim D et al (2009) Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 21:479–493
Franklin-Tong VE (1999) Signaling and modulation of pollen tube growth. Plant Cell 11:727–738
Yang ZB (1998) Signaling tip growth in plants. Curr Opin Plant Biol 1:525–530
Ryser U (1979) Cotton fiber differentiation: occurrence and distribution of coated and smooth vesicles during primary and secondary wall formation. Protoplasma 98:223–239
Ruan YL (2007) Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct Plant Biol 34:1–10
Ruan YL, Danny JL, Robert TF (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13:47–60
Lacape JM, Nguyen TB, Courtois B et al (2005) QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci 45:123–140
He DH, Lin ZX, Zhang XL et al (2007) QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica 153:181–197
Acknowledgements
This work was financially supported in part by grants from the National High-tech Program (2006AA10Z111), the National Natural Science Foundation of China (30871558), and the Program for 111 project in Ministry of Education, China (B08025).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L.K., Niu, X.W., Lv, Y.H. et al. Molecular cloning and localization of a novel cotton annexin gene expressed preferentially during fiber development. Mol Biol Rep 37, 3327–3334 (2010). https://doi.org/10.1007/s11033-009-9919-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9919-2