Abstract
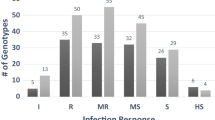
Net form of net blotch (NFNB) of barley (Hordeum vulgare L.), caused by Pyrenophora teres f. teres (Ptt) Drechsler (anamorph: Drechslera teres [Sacc.] Shoem.), is considered one of the major constraints of successful barley production in major barley growing regions of the world. Resistance to NFNB was evaluated in a barley collection of 336 genotypes (AM-2014), at seedling stage using isolates LGDPtt.19 and TD10 in the USA, and adult stage in seven hotspot environments in Morocco. The AM-2014 panel was genotyped with 9K SNP markers and genome-wide association studies (GWAS) were carried out using mixed linear model (MLM: Q + K) accounting for population structure (Q) and kinship (K) as covariates. Significant (P < 0.001) marker trait associations were corrected for false discovery rate (FDR) at the q < 0.05. Four genotypes showed an average infection response (IRs ≤ 2) to both isolates, LGDPttt.19 and TD10, at the seedling stage, and 30 genotypes showed resistance in all environments in the field while three genotypes exhibited the highest resistance at both stages. The GWAS of NFNB identified 31 distinct QTLs on all seven barley chromosomes, of which 8 with resistance at seedling stage, 21 were associated with resistance at the adult stage, and two QTLs, QRptt.2H-132.15 and QPtt.6H-54-55, conferred resistance at both stages. Of 31 resistance QTLs reported in this study, 10 QTLs coincided with previously mapped QTL while 21 are novel, thereby validating the GWAS approach used in this study. The resistance sources identified in AM-2014 and QTL mapped in this study are valuable resources for marker-assisted breeding for NFNB resistance in the future.


Similar content being viewed by others
References
Abu Qamar M, Liu ZH, Faris JD, Chao S, Edwards MC, Lai Z, Frankowiak JD, Friesen TL (2008) A region of barley chromosome 6H harbors multiple major genes associated with the net type net blotch resistance. Theor Appl Genet 117:1261–1270
Afanasenko O, Mironenko N, Filatova O, Kopahnke D, Kramer I, Ordon F (2007) Genetics of hostpathogen interactions in the Pyrenophora teres f. teres (net form) – barley (Hordeum vulgare) pathosystem. Eur J Plant Pathol 117:267–280. https://doi.org/10.1007/s10658-006-9093-5
Amezrou R, Gyawali S, Belqadi L, Chao S, Arbaoui M, Mamidi S, Rehman S, Sreedasyam A, Verma RPS (2017) Molecular and phenotypic diversity of a worldwide ICARDA spring barley collection. Genetic resources and crop evolution. Genet Resour Crop Evol:1–15
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57(1):289–300
Box GEP, Cox DR (1964) An analysis of transformations. J R Stat Soc B 26:211–252
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brown MP, Steffenson BJ, Webster RK (1993) Host range of Pyrenophora teres f. teres isolates from California. Plant Dis 77:942–974
Burlakoti RR, Gyawali S, Chao S, Smith KP, Horsley RD, Cooper B, Muehlbauer GJ, Neate SM (2016) Genome-wide association study of spot form of net blotch resistance in the Upper Midwest barley breeding programs. Phytopathology 107:100–108
Cakir M, Gupta S, Platz GJ, Ablett GA, Loughman R, Embiri LC, Poulsen D, Li C, Lance RCM, Galway NW, Jones MGK, Appels R (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f teres in barley (Hordeum vulgare L.). Aust J Agric Res 54:1369–1377
Cakir M, Gupta S, Li C, Hayden M, Mather DE, Ablett GA, Platz GJ, Broughton S, Chalmers KJ, Loughman R, Jones MGK, Lance RCM (2011) Genetic mapping and QTL analysis of disease resistance traits in the barley population Baudin x AC Metcalfe. Crop Pasture Sci 62:152–161
Caranta C, Lefebvre V, Palloix A (1997) Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol Plant-Microbe Interact 10:872–878
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Emebiri LC, Platz G, Moody DB (2005) Disease resistance genes in a doubled haploid population of two-rowed barley segregating for malting quality attributes. Aust J Agric Res 56(1):49–56
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Eyal Z, Scharen AL, Prescott JM, Van Ginkel M (1987) The Septoria diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure linkage disequilibrium in plants. Annu Rev Plan Biol 54:357–374
Friesen TL, Faris JD, Lai Z, Steffenson BJ (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a barley doubled haploid population. Genome 409:855–859
Graner A, Foroughi-Wehr B, Tekauz A (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica 91:229–234
Grewal TS, Rossnagel BG, Pozniak CJ, Scoles GJ (2008) Mapping quantitative trait loci associated with barley net blotch resistance. Theor Appl Genet 116:529–539
Hickey LT, Lawson W, Platz GJ, Dieters M, Arief VN, German S, Fletcher S, Park RF, Singh D, Pereyra S, Franckowiak J (2011) Mapping Rph20: a gene conferring adult plant resistance to Puccinia hordei in barley. Theor Appl Genet 123:55–68
Hickey LT, Lawson W, Platz GJ, Fowler RA, Arief V, Dieters M, German S, Fletcher S, Park RF, Singh D, Pereyra S, Franckowiak J (2012) Mapping quantitative trait loci for partial resistance to powdery mildew in an Australian barley population. Crop Sci 52:1021–1032
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Jalli M, Robinson J (1999) Stable resistance in barley to Pyrenophora teres f. teres isolates from the Nordic-Baltic region after increase on standard host genotypes. Euphytica 113:71–77
Jordan VW, Allen E (1984) Barley net blotch: influence of straw disposal and cultivation methods on inoculum potential, and on incidence and severity of autumn disease. Plant Pathol 33:547–559
Koladia VM, Faris JD, Richards JK, Brueggeman RS, Chao S, Friesen TL (2016) Genetic analysis of net form net blotch resistance in barley lines CIho 5791 and Tifang against a global collection of P. teres f. teres isolates. Theor Appl Genet 130(1):163–173
Konig J, Perovic D, Kopahnke D, Ordon F (2013) Development of an efficient method for assessing resistance to the net type of net blotch (Pyrenophora teres f. teres) in winter barley and mapping of quantitative trait loci for resistance. Mol Breeding 32:641–650. https://doi.org/10.1007/s11032-013-9897-x
Lai Z, Faris JD, Weiland JJ, Brian J. Steffenson BJ, Friesen TL (2007) Genetic mapping of Pyrenophora teres f. teres genes conferring avirulence on barley. Fungal Genet Biol 44:323–329
Li ZK, Arif M, Zhong DB, Fu BY, Xu JL, Domingo-Rey J, Ali J, Vijayakumar CHM, Yu SB, Khush GS (2006) Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc Natl Acad Sci 103:7994–7999
Liu Z, Ellwood SR, Oliver RP, Friesen TL (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol Plant Pathol 12(1):1–19
Ma ZQ, Lalpitan NLV, Steffenson B (2004) QTL mapping of net blotch resistance genes in a doubled-haploid population of six-rowed barley. Euphytica 137:291–296
Manninen OM, Kalendar R, Robinson J, Schulman AH (2000) Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol Genet Genomics 264:325–334
Manninen OM, Jalli M, Kalendar R, Schulman A, Afanasenko O, Robinson J (2006) Mapping of major spot-type and net-type net-blotch resistance genes in the Ethiopian barley line CI 9819. Genome 49:1564–1571
Marcel TC, Gorguet B, Ta MT, Kohutova Z, Vels A, Niks RE (2008) Isolate specificity of quantitative trait loci for partial resistance of barley to Puccinia hordei conformed in mapping populations of near-isogenic lines. New Phytol 177:743–755
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Marıa M-A, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KFX, Scholz U, Poland JA, Stein N, Waugh R (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727. https://doi.org/10.1111/tpj.12319
Mathre DE (1997) Compendium of barley diseases, 2nd edn. The American Phytopathological Society. APS Press, St. Paul
McLean MS, Howlett BJ, Turkington TK, Platz GJ, Hollaway GJ (2012) Spot form of net blotch resistance in a diverse set of barley lines in Australia and Canada. Plant Dis 96:569–576
McLean MS, Weppler R, Howlett BJ, Hollaway GJ (2016) Spot form of net blotch suppression and yield of barley in response to fungicide application in the Wimmera region of Victoria, Australia. Australas Plant Pathol 45(1):37–43
Munoz-Amatriain M, Cuesta-Marcos A, Endelman JB, Comadran J, Bonman JM et al (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS One 9(4):e94688. https://doi.org/10.1371/journal.pone.0094688
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costish DE, Buckler ES (2009) Association mapping: critical consideration shift from genotyping to experimental design. Plant Cell 21:2194–2202
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Payne RW (2013) The Guide to the GenStat ® Command Language (Release 16). Part 2: statistics. Lawes Agricultural Trust, Rothamsted Experimental Station, Harpenden
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2008) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Raman H, Platz GJ, Chalmers KJ, Raman R, Read BJ, Barr AR, Moody DB (2003) Mapping of genetic regions associated with net form of net blotch resistance in barley. Aust J Agric Res 54:1359–1367
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt WR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. PNAS 98(20):1479–11484
Richards JK, Friesen TL, Brueggeman RS (2017) Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance/susceptibility loci. Theor Appl Genet 130(5):915–927
Richter K, Schondelmaier J, Jung C (1998) Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor Appl Genet 97(8):1225–1234
Robinson J, Jalli M (1996) Diversity among Finnish net blotch isolates and resistance in barley. Euphytica 92:81–87
Roy JK, Smith KP, Muehlbauer GJ, Chao S, Close TJ, Steffenson BJ (2010) Association mapping of spot blotch resistance in wild barley. Mol Breed 26:243–256
Saari EE, Prescott JM (1975) A scale for appraising the foliar intensity of wheat disease. Plant Dis Reporter 59:377–380
Sharma RC, Duveiller E (2006) Spot blotch continues to cause substantial grain yield reductions under resource-limited farming conditions. J Phytopathol 154:482–488
Shipton WA, Khan TN, Boyd WJR (1973) Net blotch of barley. Rev Plant Pathol 52:269–290
Slotta TAB, Brady L, Chao S (2008) High throughput tissue preparation for large-scale genotyping experiments. Mol Ecol Resour 8:83–87. https://doi.org/10.1111/j.1471-8286.2007.01907.x
St. Clair DA (2010) Quantitative disease resistance and quantitative trait loci in breeding. Annu Rev Phytopathol 48:247–268
St Pierre S, Gustus C, Steffenson B, Dill-Macky R, Smith KP (2010) Mapping net form net blotch and Septoria speckled leaf blotch resistance loci in barley. Phytopathology 100(1):80–84
Steffenson BJ, Hayes HM, Kleinhofs A (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor Appl Genet 92:552–558
Steffenson B, Pederson J, Pederson V (1999) Common barley diseases in North Dakota. Extension Bulletin, NDSU Ext. Service
Tamang P, Neupane A, Mamidi S, Friesen T, Brueggeman R (2015) Association mapping of seedling resistance to spot form of net blotch in a worldwide collection of barley. Phytopathology 105:500–508
Tekauz A (1985) A numerical scale to classify reactions of barley to Pyrenophora teres. Can J Plant Pathol 7(2):181–183
Wang X, Mace ES, Platz GJ, Hunt CH, Hickey LT, Franckowiak JD, Jordan DR (2014) Spot form of net blotch resistance in barley is under complex genetic control. Theor Appl Genet 128:489–499
Weir BS (1979) Inferences about linkage disequilibrium. Biometrics 35:235–254
Wonneberger R, Ficke A, Lillemo M (2017a) Identification of quantitative trait loci associated with resistance to net form net blotch in a collection of Nordic barley germplasm. Theor Appl Genet:1–19
Wonneberger R, Ficke A, Lillemo M (2017b) Mapping of quantitative trait loci associated with resistance to net form net blotch (Pyrenophora teres f. teres) in a doubled haploid Norwegian barley population. PLoS ONE 12(4):e0175773 https://doi.org/10.1371
Young ND (1996) QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol 34:479–501
Youcef-Benkada M, Bendahmane BS, Sy AA, Barrault G, Albertini L (1994) Effects of inoculation of barley inflorescences with Drechslera teres upon location of seed-borne inoculum and its transmission of seedlings as modified by temperature and soil moisture. Plant Pathol 43:350–355
Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen BM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genet 38:203–208
Yun SJ, Gyenis L, Hayes PM, Matus I, Smith KP, Steffenson BJ, Muehlbauer GJ (2005) Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci 45(6):2563–2572
Zadok JC, Chang TT, Konzak FC (1974) A decimal code for growth stages of cereals. Weed Res 14:415–421
Zhao HH, Fernando RL, Dkeers JCM (2007) Power and precision of alternate methods for linkage disequilibrium mapping of quantitative trait loci. Genetics 175:1975–1986
Zhou H, Steffenson B (2013b) Genome-wide association mapping reveals genetic architecture of durable spot blotch resistance in US barley breeding germplasm. Mol Breed 32:139–154
Zhou H, Steffenson BJ (2013a) Association mapping of Septoria speckled leaf blotch resistance in US barley breeding germplasm. Phytopathology 103:600–609
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Ziems L, Hickey L, Hunt C, Mace E, Platz GJ, Franckowiak J, Jordan D (2014) Association mapping of resistance to Puccinia hordei in Australia barley breeding germplasm. Theor Appl Genet 127:1199–1212
Acknowledgements
This study was undertaken as part of the CGIAR Research Program on Dryland Cereals (CRP-DC). The financial supports from CRP-DC and USAID-CGIAR Linkage program are highly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amezrou, R., Verma, R.P.S., Chao, S. et al. Genome-wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol Breeding 38, 58 (2018). https://doi.org/10.1007/s11032-018-0813-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0813-2