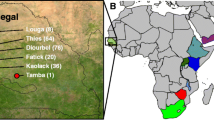
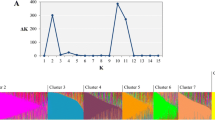
Abstract
To help support utilization of germplasm resources for tomatillo (Physalis philadelphica) crop improvement, we characterized genetic diversity in the National Plant Germplasm System collection. Genotyping by sequencing, a method of high-throughput DNA sequencing of reduced representation genomic libraries, was performed on 190 plant samples. This yielded 77,340 high-quality filtered single nucleotide polymorphisms from 179 plants sampled from 125 accessions. Geographical information systems data on geospatial references were verified using web- and PC-based software tools. We found that multiple plants sampled per accession were closely related to each other, but there was no apparent pattern related to original sampling location with respect to state in Mexico. There was no evidence for isolation by distance in a 15-accession, 53 plant geodiversity panel. Average proportion of heterozygous sites was halved in samples from nine inbred lines relative to samples from open-pollinated accessions (0.04 vs. 0.08). The genetic characterization of these accessions can help end users choose germplasm to support increased production of fresh and processed tomatillo products for expanding niche markets.

Similar content being viewed by others
Abbreviations
- AMS:
-
Agricultural Marketing Service
- BWA:
-
Burrows–Wheeler alignment
- CBSU:
-
Computational Biology Service Unit
- FAO:
-
Food and Agriculture Organization
- GBS:
-
Genotyping by sequencing
- SNP:
-
Single nucleotide polymorphism
- GIS:
-
Geographical information systems
- H e :
-
Expected heterozygosity
- SSR:
-
Simple sequence repeat
- NCBI:
-
National Center for Biotechnology Information
- NJ:
-
Neighbor joining
- NPGS:
-
National Plant Germplasm System
- SRA:
-
Sequence read archive
- PGRU:
-
Plant Genetic Resources Unit
- GRIN:
-
Germplasm Resources Information Network
- UNEAK:
-
Universal Network Enabled Analysis Kit
- USDA:
-
United States Department of Agriculture
References
Anonymous (2014) Mexico: broad and deep agricultural trade relationship. USDA Foreign Agricultural Service, Global Agricultural Information Network Report Number MX4019
Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T (2012) BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res 40(D1):D57–D63
Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39(Database issue):D1149–1155
Choi JK, Murillo G, Su BN, Pezzuto JM, Kinghorn AD, Mehta RG (2006) Ixocarpalactone A isolated from the Mexican tomatillo shows potent antiproliferative and apoptotic activity in colon cancer cells. FEBS J 273:5714–5723
Elizalde-Gonzalez MP, Hernandez-Ogarcia SG (2007) Effect of cooking processes on the contents of two bioactive carotenoids in Solanum lycopersicum tomatoes and Physalis ixocarpa and Physalis philadelphica tomatillos. Molecules 12:1829–1835
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Freeling M, Walbot V, Lee M (1994) Inbred lines of maize and their molecular markers. In: The maize handbook. Springer Lab Manuals. Springer, New York, pp 423–432
Freyre R, Loy JB (2000) Evaluation and yield trials of tomatillo in New Hampshire. HortTechnology 10:374–377
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9:e90346
Grills G, Schweitzer P, Sun Q, Pillardy J, Wang W, Stelick T, Bukowski R, Ponnala L, VanEe J, Grills G (2011) Integrated core facility support and optimization of next generation sequencing technologies. J Biomol Tech 22(Supplement):S32
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27:570–580
Howard N, Wright S (2012) Tomatillo. University of Kentucky Cooperative Extension Service. CCD Crop Profiles
Hudson WD Jr (1986) Relationships of domesticated and wild Physalis philadelphica. In: D’Arcy WG (ed) Solanaceae biology and systematics. Columbia University Press, New York, pp 416–432
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro H (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Labate JA, Robertson LD, Strickler SR, Mueller LA (2014) Genetic structure of the four wild tomato species in the Solanum peruvianum s.l. species complex. Genome 57:169–180
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Liu HY, Koike ST, Xu D, Li R (2012) First report of Turnip mosaic virus in tomatillo (Physalis philadelphica) in California. Plant Dis 96(2):296
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9(1):e1003215
Maldonado E, Perez-Castorena AL, Garces C, Martinez M (2011) Philadelphicalactones C and D and other cytotoxic compounds from Physalis philadelphica. Steroids 76(7):724–728
Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11:499–511
Melchinger AE (1990) Use of molecular markers in breeding for oligogenic disease resistance. Plant Breed 104:1–19
Menzel MY (1951) The cytotaxonomy and genetics of Physalis. Proc Am Philos Soc 95:132–183
Montes Hernández S, Aguirre Rivera J (1994) Tomatillo, husk-tomato (Physalis philadelphica). In: Hernándo Bermejo J, León J (eds) Neglected crops: 1492 from a different perspective. Plant Production and Protection Series No. 26. FAO, Rome, Italy, pp 117–122
Moriconi DN, Rush MC, Flores H (1990) Tomatillo: a potential vegetable crop for Louisiana. In: Janick J, Simon JE (eds) Advances in new crops. Timber Press, Portland, pp 407–413
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Pandey KK (1957) Genetics of self-incompatibility in Physalis ixocarpa Brot.—A new system. Am J Bot 44:879–887
Pluzhnikov A, Donnelly P (1996) Optimal sequencing strategies for surveying molecular genetic diversity. Genetics 144:1247–1262
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5:92–102
Rambaut A (2012) FigTree 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 12 May 2014
Ramirez-Godina F, Robledo-Torres V, Pournabav RF, Benavides-Mendoza A, Hernandez-Pinero JL, Reyes-Valdes MH, Alvarado-Vazquez MA (2013) Yield and fruit quality evaluation in husk tomato autotetraploids (Physalis ixocarpa) and diploids. Aust J Crop Sci 7(7):933–940
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145(4):1219–12228
Russell J, Hackett C, Hedley P, Liu H, Milne L, Bayer M, Marshall D, Jorgensen L, Gordon S, Brennan R (2014) The use of genotyping by sequencing in blackcurrant (Ribes nigrum): developing high-resolution linkage maps in species without reference genome sequences. Mol Breed 33:835–849
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S (2011) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 39(Suppl. 1):D38–D51
Simon J, Park C, Ayeni A, Payne R, Nitzsche P, Sciarappa B, Van Vranken R, Komar S, Dager E, Wu Q, Schilling B, Govindasamy R, Kelley KM (2013) Producing and marketing ethnic herbs and greens. In: Infante-Casella ML, Kline WL (eds) 58th New Jersey Agricultural Convention and Trade Show, Atlantic City, NJ, February 5–7, 2013. Rutgers Cooperative Extension, pp 140–141
Smith R, Jiminez M (1999) Tomatillo production in California. Vegetable Production Series, Pub. 7246. University of California, Division of Agriculture and Natural Resources
Su B-N, Misico R, Jung Park E, Santarsiero BD, Mesecar AD, Fong HHS, Pezzuto JM, Douglas Kinghorn A (2002) Isolation and characterization of bioactive principles of the leaves and stems of Physalis philadelphica. Tetrahedron 58(17):3453–3466
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485(7400):635–641
Upadhyaya HD, Gowda CLL (2006) Enhancing utilization of plant genetic resources in crop improvement. In: Plant breeding in post genomics era. Proceedings of Second National Plant Breeding Congress, Coimbatore, India, 1–3 March, 2006. Indian Society of Plant Breeders, pp 34–51
Vargas-Ponce O, Perez-Alvarez LF, Zamora-Tavares P, Rodriguez A (2011) Assessing genetic diversity in Mexican husk tomato species. Plant Mol Biol Rep 29(3):733–738
Vision T, Chacon M, Tsompana M, Robertson L, Pena Lomeli A, Ponce O (2006) Microsatellite variation and population structure in tomatillo (Physalis philadelphica Lam.). In: PAA/Solanaceae abstract 408, July 23–27, Madison, WI
Waterfall UT (1967) Physalis in Mexico, Central America and the West Indies. Rhodora 69:82–120, 203–239, 319–329
Wei J, Hu X, Yang J, Yang W (2012) Identification of single-copy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PLoS ONE 7(11):e50164
Willing E-M, Dreyer C, van Oosterhout C (2012) Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 7:e42649
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Zamora-Tavares P, Vargas-Ponce O, Sánchez-Martínez J, Cabrera-Toledo D (2015) Diversity and genetic structure of the husk tomato (Physalis philadelphica Lam.) in Western Mexico. Genet Resour Crop Evol 62:141–153
Acknowledgments
We thank Susan M. Sheffer, William Garman, Jonathan Spencer, Paul Kisly, Sherri Tennies and Bob Nearpass for their assistance and excellent technical support. Sharon Mitchell and Robert Bukowski (Cornell University) performed and analyzed the spike-in experiment to verify library optimization. USDA is an equal opportunity provider and employer. The use of trade, firm or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable. This research was supported by CRIS Project No. 1910-21000-024-00D. Part of this work was carried out using the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Labate, J.A., Robertson, L.D. Nucleotide diversity estimates of tomatillo (Physalis philadelphica) accessions including nine new inbred lines. Mol Breeding 35, 106 (2015). https://doi.org/10.1007/s11032-015-0302-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0302-9