Abstract
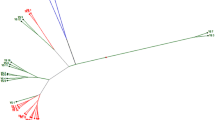
Brassica rapa is an economically important crop with a wide range of morphologies. Developing a set of fixed lines and understanding their diversity has been challenging, but facilitates resource conservation. We investigated the genetic diversity and population structure of 238 fixed lines of leafy B. rapa with 45 new simple sequence repeat markers and 109 new NGS (next-generation sequencing)-generated single nucleotide polymorphism markers evenly distributed throughout the B. rapa genome. Phylogenetic analysis classified the vegetable fixed lines into four subgroups, with the three oil types forming a separate and relatively distant cluster. A model-based population structure analysis identified four subpopulations corresponding to geographical origins and morphological traits, and revealed extensive allelic admixture. In particular, the Chinese cabbage cluster was subdivided into three groups and showed considerable correlation with leaf- and heading-related traits (leaf and heading shape). The vegetable B. rapa fixed lines successfully developed in our study could be valuable materials for establishing a multinational Brassica rapa diversity resource. Understanding the genetic diversity and population structure could be useful for utilization of the representative genetic variation and further genomic analysis.




Similar content being viewed by others
References
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polumorphisms. Am J Hum Genet 32:314–331
Cao J, Cao S, Miao Y, Lu G (1997) Cladistic operational analysis and study on the evolution of Chinese cabbage groups (Brassica campestris L.). Acta Hort Sin 24:35–42
Chen S, Nelson MN, Ghamkhar K, Fu T, Cowling WA (2008) Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51:1–10
Chen S, Wan Z, Nelson MN, Chauhan JS et al (2013) Evidence from genome-wide simple sequence repeat markers for a polyphyletic origin and secondary centers of genetic diversity of Brassica juncea in China and India. J Hered 104:416–427
Choi SR, Teakle GR, Plaha P et al (2007) The reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Theor Appl Genet 115:777–792
Delourme R, Falentin C, Forneju B et al (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genom 14:120
Duijs JG, Voorrips RE, Visser DL, Custers JBM (1992) Microspore culture is successful in most crop types of Brassica oleracea L. Euphytica 60:45–55
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ferrie AMR, Epp DJ, Keller WA (1995) Evaluation of Brassica rapa L. genotypes for microspore culture response and identification of a highly embryogenic line. Plant Cell Rep 14:580–584
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50:121–128
Gulden R, Warwick S, Thomas A (2008) The biology of Canadian weeds. 137. Brassica napus L. and B. rapa L. Can J Plant Sci 88:951–996
Guo YD, Pulli S (1996) High frequency embryogenesis in Brassica campestris microspore culture. Plant Cell Tissue Organ Cult 46:219–225
Guo Y, Chen S, Li Z, Cowling WA (2014) Centre of origin and centers of diversity in an ancient crop, Brassica rapa (turnip rape). J Hered 105:555–565
Hamblin MT, Warburton M, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 12:e1367
Harper AL, Trick M, Higgins J et al (2012) Associative transcriptomics of traits in the polyloid crop species Brassica napus. Nat Biotechnol 30:798–802
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121:475–487
Lee JG, Bonnema G, Zhang N, Kwak JH, de Vos RC, Beekwilder J (2013) Evaluation of glucosinolate variation in a collection of turnip (Brassica rapa) germplasm by the analysis of intact and desulfo glucosinolates. J Agric Food Chem 61:3984–3993
Li CW (1981a) The origin, evolution, taxonomy and hybridization of Chinese cabbage. In: Talekar NS, Griggs TD (eds) Chinese cabbage. AVRDC, Tainan, pp 3–10
Li JW (1981b) The origins and evolution of vegetable crops in China. Sci Agric Sin 14:90–95
Li X, Ramchiary N, Choi SR et al (2010) Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 53:939–947
Li X, Ramchiary N, Dhandapani V et al (2013) Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Res 20(1):1–16. doi:10.1093/dnares/dss029
Li F, Chen B, Xu K et al. (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21(4):355–367. doi:10.1093/dnares/dsu002
Lichter R (1982) Induction of haploid plants from isolated pollen of Brassica napus. Z Pflanzenphysiol 105:427–434
Liu K, Muse SV (2005) PowerMarker: an integrated analysis enviroment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu L, Qu C, Wittkop B et al (2013) A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PLoS ONE. doi:10.1371/journal.pone.0083052
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Pink D, Bailey L, McClement S et al (2008) Double haploids, markers and QTL analysis in vegetable Brassicas. Euphytica 164(2):509–514
Pino Del Carpio D, Basnet RK, De Vos RC, Maliepaard C, Paulo MJ, Bonnema G (2011a) Comparative methods for association studies: a case study on metabolite variation in a Brassica rapa core collection. PLoS ONE 6:e19624
Pino Del Carpio D, Basnet RK, De Vos RC, Maliepaard C, Visser R, Bonnema G (2011b) The patterns of population differentiation in a Brassica rapa core collection. Theor Appl Genet 6:1105–1118
Pradhan A, Nelson MN, Plummer JA, Cowling WA, Yan G (2011) Characterization of Brassica nigra collections using simple sequence repeat markers reveals distinct groups associated with geographical location, and frequent mislabelling of species identity. Genome 54:50–63
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945
Pyo HK (1981) Historical observation on the cultivar development of Chinese cabbage in Korea. In: Talekar NS, Griggs TD (eds) Chinese cabbage. AVRDC, Tainan, pp 41–47
Raman H, Dalton-Morgan J, Diffey S, Raman R, Alamery S, Edwards D, Batley J (2014) SNP markers-based map construction and genome-wide linkage analysis in Brassica napus. Plant Biotechnol J 12(7):851–860. doi:10.1111/pbi.12186
Ramchiary N, Nguyen DV, Li X et al (2011) Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res 18:305–320
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Semagn K, Magorokosho C, Ogugo V, Makumbi D, Warburton ML (2014) Genetic relationships and structure among open-pollinated maize varieties adapted to eastern and southern Africa using microsatellite markers. Mol Breeding 34(3):1423–1435. doi:10.1007/s11032-014-0126-z
Singh N, Choudhury DR, Singh AK et al (2013) Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of indian rice varieties. PLoS ONE 8:e84136
Strigens A, Schipprack W, Reif JC, Melchinger AE (2013) Unlocking the genetic diversity of maize landraces with doubled haploids opens new avenues for breeding. PLoS ONE 8:e57234
Trick M, Long Y, Meng J, Bandcroft I (2009) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using solexa transcriptome sequencing. Plant Biotechnol J 7:334–346
Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V (2014) Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theor Appl Genet 127:283–295
Wang XW, Wang HZ, Wang J et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1157
Wang W, Huang S, Liu Y et al (2012a) Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC Genom 13:523
Wang F, Li L, Li H, Liu L, Zhang Y, Gao J, Wang X (2012b) Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 99:299–307
Wong RS, Zee SY, Swanson EB (1996) Isolated microspore culture of chinese flowering cabbage (Brassica campestris ssp. parachinensis). Plant Cell Rep 15(6):396–400
Yoon JY (1982) Problems and achievements in varietal improvement and genetic studies of Chinese cabbage. Res. Report of ORD 24: sup. 450–459
Yu S, Zhang F, Wang X, Zhao X, Zhang D, Yu Y, Xu J (2010) Genetic diversity and marker-trait associations in a collection of Pak-choi (Brassica rapa L. ssp. Chinensis Makino) accessions. Genes Genomics 32:419–428
Yu X, Wang H, Zhong W, Bai J, Liu P, He Y (2013) QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PlosOne. doi:10.1371/journal.pone.0076059
Zhao J, Wang X, Deng B et al (2005) Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet 110:1301–1314
Zhao J, Artemyeva A, Pino Del Carpio D et al (2010) Design of a Brassica rapa core collection for association mapping studies. Genome 53:884–898
Acknowledgments
We thank Dr Xiaowu Wang, Deepak Pental, Takeshi Nishio, Xilin Hou, L. Visser, P. Sparrow and Thomas C. Osborn for providing materials. This work was supported by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ007992), Rural Development Administration, Republic of Korea, and The National Natural Scientific Foundation of China, Project No. 31171967.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Wenxing Pang and Xiaonan Li have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pang, W., Li, X., Choi, S.R. et al. Development of a leafy Brassica rapa fixed line collection for genetic diversity and population structure analysis. Mol Breeding 35, 54 (2015). https://doi.org/10.1007/s11032-015-0221-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0221-9