Abstract
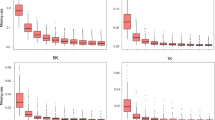
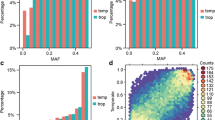
The development of single nucleotide polymorphism (SNP) markers in maize offers the opportunity to utilize DNA markers in many new areas of population genetics, gene discovery, plant breeding and germplasm identification. However, the steps from sequencing and SNP discovery to SNP marker design and validation are lengthy and expensive. Access to a set of validated SNP markers is a significant advantage to maize researchers who wish to apply SNPs in scientific inquiry. We mined 1,088 loci sequenced across 60 public inbreds that have been used in maize breeding in North America and Europe. We then selected 640 SNPs using generalized marker design criteria that enable utilization with several SNP chemistries. While SNPs were found on average every 43 bases in 1,088 maize gene sequences, SNPs that were amenable to marker design were found on average every 623 bases; representing only 7% of the total SNPs discovered. We also describe the development of a 768 marker multiplex assay for use on the Illumina® BeadArray™ platform. SNP markers were mapped on the IBM2 intermated B73 × Mo17 high resolution genetic map using either the IBM2 segregating population, or segregation in multiple parent-progeny triplets. A high degree of colinearity was found with the genetic nested association map. For each SNP presented we give information on map location, polymorphism rates in different heterotic groups and performance on the Illumina® platform.

Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Batley J, Mogg R, Edwards D, O’Sullivan H, Edwards KJ (2003) A high-throughput SNuPE assay for genotyping SNPs in flanking regions of Zea mays sequence tagged simple sequence repeats. Mol Breed 11:111–120. doi:10.1023/A:1022446021230
Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC, Tingey SV, Rafalski A (2002) Insertion-deletion polymorphisms in 3’ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol 48:539–547. doi:10.1023/A:1014841612043
Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3:19. doi:10.1186/1471-2156-3-19
Clark A, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R (2005) Ascertainment bias in studies of the human genome-wide polymorphism. Genome Res 15:1496–1502. doi:10.1101/gr.4107905
Cone K, McMullen M, Vroh Bi I, Davis G, Yim YS, Gardiner J, Polacco M, Sanchez-Villeda H, Fang Z, Schroeder S, Havermann SA, Bowers JE, Paterson AH, Soderland CA, Engler FW, Wing RA, Coe EH (2002) Genetic, physical and informatic resources for maize: on the road to an integrated map. Plant Physiol 130:1598–1605. doi:10.1104/pp.012245
Emrich SJ, Li L, Wen T-J, Yandeau-Nelson MD, Fu Y, Guo L, Chou H-H, Aluru S, Ashlock DA, Schnable PS (2007) Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175:429–439. doi:10.1534/genetics.106.064006
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Gardiner J, Schroeder SS, Polacco ML, Sanchez-Villeda H, Fang Z, Morgante M, Landewe T, Fengler K, Useche F, Hanafey M, Tingey S, Cou H, Wing R, Soderlund C, Coe EH Jr (2004) Anchoring 9371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimentional overgo hybridization. Plant Physiol 134:1317–1326. doi:10.1104/pp.103.034538
Gupta PK, Roy JK, Prasad M (2001) Single nucleotide polymorphisms: a new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr Sci 80:524–535
Jones E, Sullivan H, Bhattramakki D, Smith J (2007) A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for the genotypic analysis of maize Zea mays L. Theor Appl Genet 115:361–371. doi:10.1007/s00122-007-0570-9
Lee MN, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A (2002) Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol 48:453–461. doi:10.1023/A:1014893521186
Pettersson M, Bylund M, Alderborn A (2003) Molecular haplotype determination using allele-specific PCR and pyrosequencing technology. Genomics 82:390–396. doi:10.1016/S0888-7543(03)00177-0
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100. doi:10.1016/S1369-5266(02)00240-6
Rafalski A, Tingey S (2008) SNPs and their use in maize. In: Henry R (ed) Plant Genotyping II: SNP Technology. CAB International, Wallingford, UK, pp.30–43
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98:9161–9166. doi:10.1073/pnas.151244298
Vroh Bi I, McMullen MD, Villeda HS, Schroeder S, Gardiner J, Polacco M, Soderlund C, Wing R, Fang Z, Coe EH (2006) Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46:12–21. doi:10.2135/cropsci2004.0706
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276. doi:10.1016/0040-5809(75)90020-9
Wright SI, Vroh Bi I, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308:1310–1314. doi:10.1126/science.1107891
Yu J, Holland JB, McMullen M, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genet 178:539–551. doi:10.1534/genetics.107.074245
Zhao W, Canaran P, Jurkuta R, Fulton T, Glaubitz J, Buckler E, Doebley J, Gaut B, Goodman M, Holland J, Kresovich S, McMullen M, Stein L, Ware D (2006) Panzea: a database and resource for molecular and functional diversity in the maize genome. Nucleic Acids Res 34:D725–D757. doi:10.1093/nar/gkl196
Zwick ME, Cutler DJ, Chakavarti A (2000) Patterns if genetic variation in Mendelian and complex traits. Annu Rev Genomics Hum Genet 1:387–407. doi:10.1146/annurev.genom.1.1.387
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jones, E., Chu, WC., Ayele, M. et al. Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol Breeding 24, 165–176 (2009). https://doi.org/10.1007/s11032-009-9281-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9281-z