Abstract
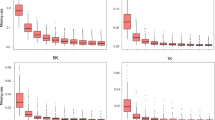
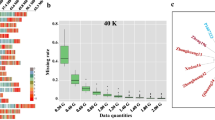
We report on the comparative utilities of simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers for characterizing maize germplasm in terms of their informativeness, levels of missing data, repeatability and the ability to detect expected alleles in hybrids and DNA pools. Two different SNP chemistries were compared; single-base extension detected by Sequenom MassARRAY®, and invasive cleavage detected by Invader® chemistry with PCR. A total of 58 maize inbreds and four hybrids were genotyped with 80 SSR markers, 69 Invader SNP markers and 118 MassARRAY SNP markers, with 64 SNP loci being common to the two SNP marker chemistries. Average expected heterozygosity values were 0.62 for SSRs, 0.43 for SNPs (pre-selected for their high level of polymorphism) and 0.63 for the underlying sequence haplotypes. All individual SNP markers within the same set of sequences had an average expected heterozygosity value of 0.26. SNP marker data had more than a fourfold lower level of missing data (2.1–3.1%) compared with SSRs (13.8%). Data repeatability was higher for SNPs (98.1% for MassARRAY SNPs and 99.3% for Invader) than for SSRs (91.7%). Parental alleles were observed in hybrid genotypes in 97.0% of the cases for MassARRAY SNPs, 95.5% for Invader SNPs and 81.9% for SSRs. In pooled samples with mixtures of alleles, SSRs, MassARRAY SNPs and Invader SNPs were equally capable of detecting alleles at mid to high frequencies. However, at low frequencies, alleles were least likely to be detected using Invader SNP markers, and this technology had the highest level of missing data. Collectively, these results showed that SNP technologies can provide increased marker data quality and quantity compared with SSRs. The relative loss in polymorphism compared with SSRs can be compensated by increasing SNP numbers and by using SNP haplotypes. Determining the most appropriate SNP chemistry will be dependent upon matching the technical features of the method within the context of application, particularly in consideration of whether genotypic samples will be pooled or assayed individually.




Similar content being viewed by others
References
Batley J, Mogg R, Edwards D, O’Sullivan H, Edwards KJ (2003a) A high-throughput SNuPE assay for genotyping SNPs in flanking regions of Zea mays sequence tagged simple sequence repeats. Mol Breed 11:111–120
Batley J, Barker G, O’Sullivan H, Edwards KJ, Edwards D (2003b) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 132:84–91
Bernardo R, Romero-Severson J, Ziegle J, Hauser J, Joe L, Hookstra G, Doerge RW (2000) Parental contribution and coefficient of coancestry among maize inbreds: pedigree, RFLP, and SSR data. Theor Appl Genet 100:552–556
Berry DA, Seltzer JD, Xie C, Wright DL, Smith JSC (2002) Assessing probability of ancestry using simple sequence repeat profiles: applications to maize hybrids and inbreds. Genetics 161:813–824
Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC, Tingey SV, Rafalski A (2002). Insertion–deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol 48:539–547
Bovo D, Rugge M, Shiao YH (1998) Origin of spurious multiple bands in the amplification of microsatellite sequences. Mol Pathol 52:50–51
Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3:19
Cone K, McMullen M, Vroh Bi I, Davis G, Yim YS, Gardiner J, Polacco M, Sanchez-Villeda H, Fang Z, Schroeder S, Havermann SA, Bowers JE, Paterson AH, Soderland CA, Engler FW, Wing RA, Coe EH (2002) Genetic, physical and informatic resources for maize: on the road to an integrated map. Plant Physiol 130:1598–1605
Cordeiro G, Eliott F, McIntyre CL, Casu RE, Henry RJ (2006) Characterization of single nucleotide polymorphisms in sugarcane ESTs. Theor Appl Genet 113:331–343
Cui Z, Carter TE Jr, Burton JW (2000) Genetic diversity patterns in Chinese soybean cultivars based on coefficient of parentage. Crop Sci 40:1780–1793
Davison A, Chilba S (2003) Laboratory temperature variation is a previously unrecognized source of genotyping error during capillary electrophoresis. Mol Ecol Notes 3:321–323
Dubreuil P, Charcosset A (1998) Genetic diversity within and among maize populations: a comparison between isozyme and nuclear RFLP loci. Theor Appl Genet 96:577–587
Estoup A, Tailliez C, Cornuet JM, Solignac M (1995) Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae). Mol Biol Evol 12:1074–1084
Fernando P, Evans BJ, Morales JC, Melnick DJ (2001) Electrophoresis artifacts—a previously unrecognized cause of error in microsatellite analysis. Mol Ecol Notes 1:235–328
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CL Jr, de Souza AP (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Gardiner J, Schroeder SS, Polacco ML, Sanchez-Villeda H, Fang Z, Morgante M, Landewe T, Fengler K, Useche F, Hanafey M, Tingey S, Cou H, Wing R, Soderlund C, Coe EH Jr (2004) Anchoring 9371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol 134:1317–1326
George MLC, Regalado E, Li W, Cao M, Dahlan M, Pabendon M, Warburton ML, Xianchun X, Hoisington D (2004) Molecular characterization of Asian maize inbred lines by multiple laboratories. Theor Appl Genet 109:80–91
Giancola S, McKhann HI, Berard A, Camilleri C, Durand S, Libeau P, Roux F, Rebound X, Gut IG, Brunel D (2006) Utilization of three high-throughput SNP genotyping methods, the GOOD assay, Amplifluor and Taqman, in diploid and polyploidy plants. Theor Appl Genet 112:1115–1124
Gizlice JAA, Carter TE, Burton J (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143–1151
Gupta PK, Roy JK, Prasad M (2001) Single nucleotide polymorphisms: a new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr Sci 80:524–535
Hatcher SL, Lambert QT, Raymond LT, Carlson JR (1993) Heteroduplex formation: a potential source of errors from PCR products. Prenat Diagn 13:171–177
Heckenberger M, Bohn M, Ziegle JS, Joe LK, Hauser JD, Hutton M, Melchinger AE (2002) Variation of DNA fingerprints among accessions within maize inbred lines and implications for identification of essentially derived varieties: I genetic and technical sources of variation in SSR data. Mol Breed 10:181–191
Heckenberger M, van der Voort JR, Melcinger AE, Peleman J, Bohn M (2003) Variation of DNA fingerprints among accessions within maize inbred lines and implications for identification of essentially derived varieties: II genetic and technical sources of variation in AFLP data and comparison with SSR data. Mol Breed 12:97–106
Isibashi Y, Saitoh T, Abe S, Yoshida MC (1996) Null microsatellite alleles due to nucleotide sequence variation in the grey-sided vole. Mol Ecol 5:589–590
Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129:440–450
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linacero R, Vasquez A, Karp A (1997). Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3:381–390
Lahermo P, Liljedahl U, Alnaes G, Axelsson T, Borrkes A, Ellonen P, Groop P, Halldén C, Holmberg D, Holmberg K, Keinanän M, Kepp K, Kere J, Kiviluoma P, Kristensen V, Lindgren C, Odeberg J, Osterman P, Parkkonen M, Saarela J, Sterner M, Strömqvist L, Talas U, Wessman M, Palotie A, Syvänen A (2006) A quality assessment survey of SNP genotyping laboratories. Hum Mutat 27:711–714
Lu H, Bernardo R (2001) Molecular marker diversity among current and historical maize inbreds. Theor Appl Genet 103:613–617
Lübberstedt T, Melchinger AE, Duble C, Vuylsteke M, Kuiper M (2000) Relationships among early European maize inbreds: IV genetic diversity revealed with AFLP markers and comparison with RFLP, RAPD, and pedigree data. Crop Sci 40:783–791
Malécot G (1948) Les mathématiques de l’hérédité. Masson and Cie, Paris. [English translation. The mathematics of heredity (1969). W.H. Freeman and Co., San Francisco]
Nei M, Li W (1979) Mathematical model for studying genetic variance in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nei M, Li W (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 106–107
Pati N, Schwinsky V, Kokanovic O, Magnuson V, Ghosh S (2004) A comparison between SNaPshot, pyrosequencing and biplex invader SNP genotyping methods: accuracy, cost and throughput. J Biochem Biophys Methods 60:1–12
Pejic I, Ajmone-Marsan P, Morgante M, Kosumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSR, and AFLPs. Theor Appl Genet 97:1248–1255
Rafalski JA (2002) Novel genetic mapping tools in plants: SNPs and LD-based approaches. Plant Sci 162:329–333
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms I barley: Mendelian inheritance, chromosomal locations, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Taramino G, Tingey S (1996) Simple sequence repeats for germplasm analysis and mapping in maize. Genome 39:277–287
Tenaillon MI, Sawkins MC, Long AD, aut RL, Doebley JF, gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L). PNAS 98:9161–9166
Vroh Bi I, McMullen MD, Villeda HS, Schroeder S, Gardiner J, Polacco M, Soderlund C, Wing R, Fang Z, Coe EH (2006) Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46:12–21
Wang Z, Weber JL, Zhong G, Tanksley SD (1994) Survey of plant short tandem DNA repeats. Theor Appl Genet 88:1–6
Warburton ML, Xianchun X, Crossa J, Franco J, Melchinger AE, Frisch M, Bohn M, Hoisington D (2002) Genetic characterization of CIMMYT inbred maize lines and open pollinated populations using large scale fingerprinting methods. Crop Sci 42:1832–1840
Werner M, Sych M, Herbon N, Illig T, Konig IR, Wjst M (2002) Large-scale determination of SNP allele frequencies in DNA pools using MALDI-TOF mass spectrometry. Hum Mut 20:57–64
Zhao W, Canaran P, Jurkuta R, Fulton T, Glaubitz J, Buckler E, Doebley J, Gaut B, Goodman M, Holland J, Kresovich S, McMullan M, Stein L, Ware D (2006) Panzea: a database and resource for molecular and functional diversity in the maize genome. Nucleic Acids Res 34:752–757
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jones, E.S., Sullivan, H., Bhattramakki, D. et al. A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for the genotypic analysis of maize (Zea mays L.). Theor Appl Genet 115, 361–371 (2007). https://doi.org/10.1007/s00122-007-0570-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0570-9