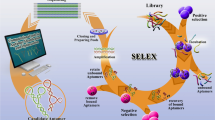
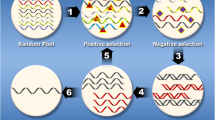
Abstract
Today, RNA aptamers are being considered promising theranostic tools against a wide variety of disorders. RNA aptamers can fold into complex shapes and bind to diverse nanostructures, macromolecules, cells, and viruses. It is possible to isolate RNA aptamers from a vast pool of nucleic acids via the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) method. As therapeutics, aptamers have great potential because of their ability to bind to proteins and selectively limit their activities with negligible side effects. Several RNA aptamers with potential implications in cancer diagnosis are known to confer a great affinity for single-stranded DNA molecules, long non-coding RNAs, circulating tumor cells, vascular endothelial growth factors, and tissue and sera-derived exosomes in patients with different malignancies. Furthermore, clinical investigations have revealed the efficacy of RNA aptamer-based agents in imaging modalities. This review seeks to deliver new insights into the development, classification, nanomerization, and modification of RNA aptamers, as well as their applications in cancer theranostics. The aptamers’ mechanism of action and their interest to clinical trials as theranostic agents are also discussed.
Graphical Abstract







Similar content being viewed by others
Data availability
This article's data sharing is not applicable as no new data were created or analyzed in this study.
References
Hathout L et al (2021) A multi-institutional analysis of adjuvant chemotherapy and radiation sequence in women with stage IIIC endometrial cancer. IntJ Radiat Oncol Biol Phys 110(5):1423–1431
Shirvalilou S et al (2021) Magnetic Hyperthermia as an adjuvant cancer therapy in combination with radiotherapy versus radiotherapy alone for recurrent/progressive glioblastoma: A systematic review. J Neurooncol 152(3):419–428
Zeng R, Dong J (2021) The Hippo signaling pathway in drug resistance in cancer. Cancers 13(2):318
Irajirad R et al (2019) Combined thermo-chemotherapy of cancer using 1 MHz ultrasound waves and a cisplatin-loaded sonosensitizing nanoplatform: An in vivo study. Cancer Chemother Pharmacol 84(6):1315–1321
Battogtokh G et al (2018) Mitochondrial-targeting anticancer agent conjugates and nanocarrier systems for cancer treatment. Front Pharmacol 9:922
Subjakova V, Oravczova V, Hianik T (2021) Polymer Nanoparticles and Nanomotors Modified by DNA/RNA Aptamers and Antibodies in Targeted Therapy of Cancer. Polymers 13(3):341
Qi S et al (2022) Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment. Biotechnol Adv 55:107902
Narayan C, Veeramani S, Thiel WH (2022) Optimization of RNA aptamer SELEX methods: improved aptamer transcript 3′-end homogeneity, PAGE purification yield, and target-bound aptamer RNA recovery. Nucleic Acid Ther 32(1):74–80
Devi S et al (2021) Aptamer-based diagnostic and therapeutic approaches in animals: Current potential and challenges. Saudi J Biological Sci 28(9):5081–5093
Ganesh K, Massagué J (2021) Targeting metastatic cancer. Nat Med 27(1):34–44
Li, Z., et al., Advances in screening and development of therapeutic aptamers against cancer cells. Frontiers in Cell and Developmental Biology, 9:2021.
Ștefan G et al (2021) Aptamers in biomedicine: Selection strategies and recent advances. Electrochim Acta 376:137994
Yan AC, Levy M (2009) Aptamers and aptamer targeted delivery. RNA Biol 6(3):316–320
Boussebayle A, Groher F, Suess B (2019) RNA-based capture-SELEX for the selection of small molecule-binding aptamers. Methods 161:10–15
Wang T et al (2019) Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol Adv 37(1):28–50
Harbaugh SV et al (2018) Screening and selection of artificial riboswitches. Methods 143:77–89
Wei X et al (2022) A review: Construction of aptamer screening methods based on improving the screening rate of key steps. Talanta 253:124003
Adachi T, Nakamura Y (2019) Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 24(23):4229
Kohlberger M, Gadermaier G (2021) SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol Appl Biochem 69:1771
Sefah K et al (2010) Development of DNA aptamers using Cell-SELEX. Nat Protoc 5(6):1169–1185
Germer K, Leonard M, Zhang X (2013) RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol 4(1):27
Thiel WH et al (2015) Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. RNA Interference. Springer, pp 187–199
Chen C et al (2017) Nucleic acid aptamer application in diagnosis and therapy of colorectal cancer based on cell-SELEX technology. NPJ precision oncol 1(1):1–7
Zhou J, Rossi J (2017) Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Disc 16(3):181–202
Cheng C et al (2013) In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol Ther Nucleic Acids 2:e67
Gopinath SCB (2007) Methods developed for SELEX. Anal Bioanal Chem 387(1):171–182
Kang K, Lee Y, Zhong J (2013) Future Trends in Biotechnology.
Lou X et al (2009) Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci 106(9):2989–2994
Ozer A, Pagano JM, Lis JT (2014) New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol Ther Nucleic Acids 3:e183
Bayat P et al (2018) SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 154:132–155
Amano R et al (2021) Specific inhibition of FGF5-induced cell proliferation by RNA aptamers. Sci Rep 11(1):1–9
Wang H et al (2018) In vivo SELEX of an inhibitory NSCLC-specific RNA aptamer from PEGylated RNA library. Mol Ther Nucleic Acids 10:187–198
Wang H et al (2019) Characterization of a bifunctional synthetic RNA aptamer and a truncated form for ability to inhibit growth of non-small cell lung cancer. Sci Rep 9(1):1–12
Dassie JP et al (2014) Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 22(11):1910–1922
Lupold SE et al (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Can Res 62(14):4029–4033
Lupold SE et al (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 62(14):4029–4033
Ul-Haq A et al (2019) Isolation of MLL1 Inhibitory RNA Aptamers. Biomolecules & therapeutics 27(2):201
Nuzzo S et al (2020) The role of RNA and DNA aptamers in glioblastoma diagnosis and therapy: a systematic review of the literature. Cancers 12(8):2173
Affinito A et al (2020) Targeting ephrin receptor tyrosine kinase A2 with a selective aptamer for glioblastoma stem cells. Mol Ther Nucleic Acids 20:176–185
Hu Q et al (2018) DNA nanotechnology-enabled drug delivery systems. Chem Rev 119(10):6459–6506
Zhou J et al (2012) Current progress of RNA aptamer-based therapeutics. Front Genet 3:234
Morita Y et al (2018) Aptamer therapeutics in cancer: current and future. Cancers 10(3):80
Sheervalilou R et al (2020) Electrochemical nano-biosensors as novel approach for the detection of lung cancer-related MicroRNAs. Curr Mol Med 20(1):13–35
Sheervalilou R et al (2019) Circulating MiR-10b, MiR-1 and MiR-30a expression profiles in lung cancer: possible correlation with clinico-pathologic characteristics and lung cancer detection. Int J Mol Cell Med 8(2):118
Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discovery 12(11):847–865
Reif, R., Guo P. (2013) Design and Construction of RNA Nanoparticles Targeting Prostate Cancer. RNA Nanotechnology and Therapeutics, p. 389.
Đapić V et al (2003) Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res 31(8):2097–2107
Mann MJ, Conte MS (2003) Transcription factor decoys for the prevention of vein bypass graft failure. Am J Cardiovasc Drugs 3(2):79–85
Sullenger BA et al (1990) Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 63(3):601–608
Kohn DB et al (1999) A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34+ cells from the bone marrow of human immunodeficiency virus-1–infected children. Blood, J Am Soc Hematol 94(1):368–371
DiGiusto DL et al (2010) RNA-based gene therapy for HIV with lentiviral vector–modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Science Translat Med 2(36):36ra43
Li M-J et al (2005) Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 12(5):900–909
Porciani D et al (2015) Aptamer-Mediated Codelivery of Doxorubicin and NF-κB Decoy Enhances Chemosensitivity of Pancreatic Tumor Cells. Molecular Therapy - Nucleic Acids 4:e235
Sullenger BA, White RR, Rusconi CP, Aptamers T, Antidotes: A Novel Approach to Safer Drug Design. (2003) Berlin. Springer, Berlin Heidelberg, Heidelberg
Rusconi CP et al (2004) Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol 22(11):1423–1428
Rusconi CP et al (2002) RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419(6902):90–94
Chan MY et al (2008) Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation 117(22):2865–2874
Povsic TJ, et al. (2011) A randomized, partially blinded, multicenter, active-controlled, dose-ranging study assessing the safety, efficacy, and pharmacodynamics of the REG1 anticoagulation system in patients with acute coronary syndromes: design and rationale of the RADAR Phase IIb trial. Am Heart J 161(2): 261–268.e2.
Heckel A, Mayer G (2005) Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J Am Chem Soc 127(3):822–823
Vorobyeva M, Vorobjev P, Venyaminova A (2016) Multivalent aptamers: Versatile tools for diagnostic and therapeutic applications. Molecules 21(12):1613
Omer M et al (2020) Improved cancer targeting by multimerizing aptamers on nanoscaffolds. Mol Ther Nucleic Acids 22:994–1003
McNamara JO et al (2008) Multivalent 4–1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Investig 118(1):376–386
Pratico ED, Sullenger BA, Nair SK (2013) Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther 23(1):35–43
Jeong H et al (2017) Multivalent Aptamer–RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells. Macromol Biosci 17(4):1600343
Bai C et al (2020) Self-Assembled Multivalent Aptamer Nanoparticles with Potential CAR-like Characteristics Could Activate T Cells and Inhibit Melanoma Growth. Mol Ther Oncol 17:9–20
Mammen M, Choi SK, Whitesides GM (1998) Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed 37(20):2754–2794
Mallik PK et al (2010) Commandeering a biological pathway using aptamer-derived molecular adaptors. Nucleic Acids Res 38(7):e93–e93
Dupont DM et al (2016) Building a molecular trap for a serine protease from aptamer and peptide modules. Bioconjug Chem 27(4):918–926
Wen J et al (2022) Ternary electrochemiluminescence biosensor based on black phosphorus quantum dots doped perylene derivative and metal organic frameworks as a coreaction accelerator for the detection of chloramphenicol. Microchem J 172:106927
Thiel KW, Giangrande PH (2009) Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 19(3):209–222
Ni S et al (2021) Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces 13(8):9500–9519
Blank M, Blind M (2005) Aptamers as tools for target validation. Curr Opin Chem Biol 9(4):336–342
Kiire CA et al (2015) Intravitreal pegaptanib for the treatment of ischemic diabetic macular edema. Clin Ophthalmol (Auckland, NZ) 9:2305
Biesecker G et al (1999) Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 42(1–3):219–230
Lin Y et al (1996) High-affinity and specific recognition of human thyroid stimulating hormone (hTSH) by in vitro-selected 2′-amino-modified RNA. Nucleic Acids Res 24(17):3407–3414
Burmeister PE et al (2006) 2-Deoxy purine, 2-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 16(4):337–351
Ng EW et al (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discovery 5(2):123–132
Becker RC, Rusconi C, Sullenger B (2005) Nucleic acid aptamers in therapeutic anticoagulation. Thromb Haemost 93(06):1014–1020
Kang K-N, Lee Y-S (2012) RNA aptamers: a review of recent trends and applications. Future Trends in Biotechnology, 153–169.
Ni X et al (2011) Nucleic acid aptamers: clinical applications and promising new horizons. Curr Med Chem 18(27):4206–4214
Ashley GW (1992) Modeling, synthesis, and hybridization properties of (L)-ribonucleic acid. J Am Chem Soc 114(25):9731–9736
Nolte A et al (1996) Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat Biotechnol 14(9):1116–1119
Fearon DT (2014) The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol Res 2(3):187–193
Suarez-Carmona M et al (2021) Combined inhibition of CXCL12 and PD-1 in MSS colorectal and pancreatic cancer: modulation of the microenvironment and clinical effects. J Immunother Cancer 9(10):e002505
Carr MW et al (1994) Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc Natl Acad Sci 91(9):3652–3656
Ninichuk V et al (2008) Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36–3′ PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am J Pathol 172(3):628–637
Bartneck M et al (2019) The CCR2+ macrophage subset promotes pathogenic angiogenesis for tumor vascularization in fibrotic livers. Cell Mol Gastroenterol Hepatol 7(2):371–390
Cai S et al (2018) Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 143(22):5317–5338
Ochoa S, Milam VT (2020) Modified nucleic acids: Expanding the capabilities of functional oligonucleotides. Molecules 25(20):4659
Debnath M, Prasad GB, Bisen PS (2010) Molecular diagnostics: promises and possibilities. Springer Science & Business Media, Berlin
Levin AA, Rosie ZY, Geary RS (2007) Basic principles of the pharmacokinetics of antisense oligonucleotide drugs. Antisense drug technology. CRC Press, pp 201–234
Kovacevic KD, Gilbert JC, Jilma B (2018) Pharmacokinetics, pharmacodynamics and safety of aptamers. Adv Drug Deliv Rev 134:36–50
Spiel AO et al (2009) The aptamer ARC1779 is a potent and specific inhibitor of von Willebrand Factor mediated ex vivo platelet function in acute myocardial infarction. Platelets 20(5):334–340
Ni S et al (2017) Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int J Mol Sci 18(8):1683
Mann AP et al (2011) Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget 2(4):298
Dougan H et al (2000) Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol 27(3):289–297
Jilma-Stohlawetz P et al (2011) A dose ranging phase I/II trial of the von Willebrand factor inhibiting aptamer ARC1779 in patients with congenital thrombotic thrombo-cytopenic purpura. Thromb Haemost 106(09):391–397
Gilbert JC et al (2007) First-in-human evaluation of anti–von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 116(23):2678–2686
Siller-Matula JM et al (2012) ARC15105 is a potent antagonist of von Willebrand factor mediated platelet activation and adhesion. Arterioscler Thromb Vasc Biol 32(4):902–909
Povsic TJ et al (2011) Pegnivacogin results in near complete FIX inhibition in acute coronary syndrome patients: RADAR pharmacokinetic and pharmacodynamic substudy. Eur Heart J 32(19):2412–2419
Phillips JA et al (2008) Applications of aptamers in cancer cell biology. Anal Chim Acta 621(2):101–108
Ulrich H (2006) RNA aptamers: from basic science towards therapy. RNA towards Medicine, p. 305–326.
Marangoni K et al (2015) Prostate-specific RNA aptamer: promising nucleic acid antibody-like cancer detection. Sci Rep 5(1):1–13
Esposito CL et al (2021) Identification of a novel RNA aptamer that selectively targets breast cancer exosomes. Mol Ther Nucleic Acids 23:982–994
Cho H et al (2012) Single-step nanoplasmonic VEGF165 aptasensor for early cancer diagnosis. ACS Nano 6:7607–7614
Safarpour H et al (2020) Optical and electrochemical-based nano-aptasensing approaches for the detection of circulating tumor cells (CTCs). Biosens Bioelectron 148:111833
Dua P et al (2013) Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Can Res 73(6):1934–1945
Li Q et al (2020) Aptamers: a novel targeted theranostic platform for pancreatic ductal adenocarcinoma. Radiat Oncol 15(1):1–12
Ray P, Sullenger BA, White RR (2013) Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: cyclophilin B and its posttranslational modifications. Nucleic Acid Ther 23(6):435–442
Hu M, Zhang K (2013) The application of aptamers in cancer research: an up-to-date review. Future Oncol 9(3):369–376
Kong RM et al (2011) Aptamer-assembled nanomaterials for biosensing and biomedical applications. Small 7(17):2428–2436
Wang AZ et al (2008) Superparamagnetic iron oxide nanoparticle–aptamer bioconjugates for combined prostate cancer imaging and therapy. Chem Med Chem: Chemistry Enabling Drug Discovery 3(9):1311–1315
Bagalkot V et al (2007) Quantum dot− aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett 7(10):3065–3070
Kim D, Jeong YY, Jon S (2010) A drug-loaded aptamer− gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano 4(7):3689–3696
Kang WJ et al (2009) Multiplex imaging of single tumor cells using quantum-dot-conjugated aptamers. Small 5(22):2519–2522
Wang AZ, Farokhzad OC (2014) Current progress of aptamer-based molecular imaging. J Nucl Med 55(3):353–356
Gold L et al. (2010) Aptamer-based multiplexed proteomic technology for biomarker discovery. Nature Precedings, 2010: p. 1–1.
Ray P et al (2012) Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J Clin Investig 122(5):1734–1741
Tong X et al (2022) Progress in cancer drug delivery based on AS1411 oriented nanomaterials. J Nanobiotechnol 20(1):1–36
Christian S et al (2003) Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J Cell Biol 163(4):871–878
Kaur H et al (2018) Aptamers in the therapeutics and diagnostics pipelines. Theranostics 8(15):4016
Xing H et al (2013) Selective delivery of an anticancer drug with aptamer-functionalized liposomes to breast cancer cells in vitro and in vivo. J Mater Chem B 1(39):5288–5297
Baek SE et al (2014) RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J Control Release 196:234–242
Zhao N et al (2013) An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small 9(20):3477–3484
Chen D et al (2016) Dual targeting luminescent gold nanoclusters for tumor imaging and deep tissue therapy. Biomaterials 100:1–16
Shiao Y-S et al (2014) Aptamer-functionalized gold nanoparticles as photoresponsive nanoplatform for co-drug delivery. ACS Appl Mater Interfaces 6(24):21832–21841
Hori S-I et al (2018) Current advances in aptamers for cancer diagnosis and therapy. Cancers 10(1):9
Kim Y-H et al (2011) An RNA aptamer that specifically binds pancreatic adenocarcinoma up-regulated factor inhibits migration and growth of pancreatic cancer cells. Cancer Lett 313(1):76–83
Kryza D et al (2016) Ex vivo and in vivo imaging and biodistribution of aptamers targeting the human matrix metalloprotease-9 in melanomas. PLoS ONE 11(2):e0149387
Affinito A et al (2019) The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol Ther Nucleic Acids 18:99–109
Fechter P et al (2019) RNA aptamers targeting integrin α5β1 as probes for cyto-and histofluorescence in Glioblastoma. Mol Ther Nucleic Acids 17:63–77
Ibarra LE et al (2022) Selective photo-assisted eradication of Triple-Negative breast cancer cells through aptamer decoration of doped conjugated polymer nanoparticles. Pharmaceutics 14(3):626
Sundaram P et al (2013) Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 48(1–2):259–271
Friberg TR, Tolentino M, L.S. Group (2010) Pegaptanib sodium as maintenance therapy in neovascular age-related macular degeneration: the LEVEL study. Br J Ophthalmol 94(12):1611–1617
Sultan MB et al (2011) A phase 2/3, multicenter, randomized, double-masked, 2-year trial of pegaptanib sodium for the treatment of diabetic macular edema. Ophthalmology 118(6):1107–1118
Noxxon Pharma, 2010b. Noxxon Initiates Multiple Dose Phase I Clinical Trail of SDF-1i nhibitor NOX-A12. <http://www.noxxon.com/downloads/pressrel/2010-09-22NOXXON_INITIATE_MULTIPLE_DOSE_PHASE_I_CLINICAL_TRIAL_OF_SDF-1_INHIBITOR_NOX-A12.pdf> (accessed 03.07.12).
Retina Today Archive, Cousins SW, 2009b. Intravitreal Anti-VEGF and Anti-PDGF Combination Therapy. <http://bmctoday.net/retinatoday/2009/10/article.asp?f=1009_12.php> (accessed 05.07.12).
Zarbin MA, Rosenfeld PJ (2010) Pathway-based therapies for age-related macular degeneration: an integrated survey of emerging treatment alternatives. Retina 30(9):1350–1367
Ophthotech, 2012b. Ophthotech’s Novel Anti-PDGF Combination Agent Fovista™Demonstrated Superior Efficacy over Lucentis Monotherapy in Large Controlled Wet AMD Trial. <http://www.ophthotech.com/ophthotechs-antipdgf-fovista-superior-efficacy-phase2b/> (accessed 03.07.12).
Ricklin D, Lambris JD (2007) Complement-targeted therapeutics. Nat Biotechnol 25(11):1265–1275
Vavalle J et al (2012) A phase 1 ascending dose study of a subcutaneously administered factor IXa inhibitor and its active control agent. J Thromb Haemost 10(7):1303–1311
Hicke BJ et al (2001) Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem 276(52):48644–48654
Keefe AD, Pai S, Ellington A (2010) Aptamers as therapeutics. Nat Rev Drug Discovery 9(7):537–550
Gilbert GE, Mast AE (2011) Curbing an inhibitor for hemostasis. Blood, J Am Soc Hematol 117(20):5277–5278
Waters EK et al (2011) Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood, J Am Soc Hematol 117(20):5514–5522
Waters E et al (2009) Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole blood. J Thromb Haemost 7:683
Fierce Healthcare, 2008. Nuvelo Announces Positive Proof-of-Concept Data With Anticogulant NU172. http://www.fiercehealthcare.com/press-releases/nuvelo-announces-positive-proof-concept-data-anticogulant-nu172(accessed 09.06.12).
Rosenberg J et al. (2010) A phase II, single-arm study of AS1411 in metastatic renal cell carcinoma (RCC). J Clin Oncol 28(15_suppl): 4590–4590.
Bates PJ et al (2009) Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 86(3):151–164
Laber D et al. (2006) Extended phase I study of AS1411 in renal and non-small cell lung cancers. J Clin Oncol 24(18_suppl): 13098–13098.
Kim MY, Jeong S (2011) In vitro selection of RNA aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther 21(3):173–178
Noxxon Pharma, 2010a. NOXXON Announces the Completion of the First-in-Human Clinical Trial with Spiegelmer NOX-A12. http://www.noxxon.com/downloads/pressrel/100503_NOXXON_Announces_the_Completion_of_theFirst-in-Human_Clinical_Trial_with_Spiegelmer_NOX-A_12.pdf (accessed 04.07.12).
NIH, 2009a. NOX-A12 First-in-Human (FIH) Study. http://www.clinicaltrials.gov/ct2/show/NCT00976378 (accessed 04.01.12).
PRISMA-PET - Primary Staging of Prostate Cancer With PSMA. https://clinicaltrials.gov/ct2/show/NCT05123300?term=PSMA&cond=Prostate+Cancer&draw=2&rank=4, 2021.
PD1 Integrated Anti-PSMA CART in Treating Patients With Castrate-Resistant Prostate Cancer. https://clinicaltrials.gov/ct2/show/NCT04768608?term=PSMA&cond=Prostate+Cancer&draw=2&rank=6, 2021-2022.
Li N et al (2011) Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One 6(6):e20299
Cerchia L et al (2012) Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther 20(12):2291–2303
Noxxon Pharma, 2009. Noxxon Announces Initiation of First Clinical Trial with a Spiegelmer http://www.noxxon.com/downloads/pressrel/090608NOXXON_Announces_Initiation_of_First_Clinical_Trial_with_a_Spiegelmer.pdf (accessed 04.07.12).
Noxxon Pharma, 2012. NOXXON Initiates Phase IIa of Anti-CCL2/MCP-1 Spiegelmer NOX-E36 for Treatment of Diabetic Nephropathy. http://www.noxxon.com/downloads/pressrel/2012-06-19_NOXXON_Initiates_Phase_IIa_of_Spiegelmer_NOX-E36.pdf (accessed 03.07.12).
Author information
Authors and Affiliations
Contributions
Conceptualization, SS; writing-original draft preparation, MR, SJ, RA, RS, SS, and SM; writing-review and editing, SS, AR, MS, SF-K, SP; supervision, SS, RS, SP. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that there is no conflict of interest regarding the publication of this article.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Razlansari, M., Jafarinejad, S., rahdar, A. et al. Development and classification of RNA aptamers for therapeutic purposes: an updated review with emphasis on cancer. Mol Cell Biochem 478, 1573–1598 (2023). https://doi.org/10.1007/s11010-022-04614-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04614-x