Abstract
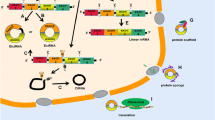
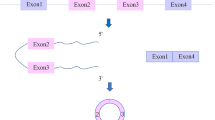
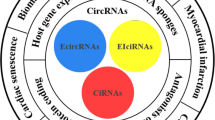
Circular RNAs (circRNAs), a novel class of endogenous noncoding RNA, are characterized by their covalently closed-loop structures without a 5′ cap or a 3′ poly(A) tail. With the evolution of high-throughput sequencing technology and bioinformatics, an increasing number of circRNAs have been discovered, and their functions were highlighted. Cardiovascular diseases (CVDs) have become the world’s leading killers, with serious impacts on human health. Although significant progress has been made in clarifying the development of CVDs from the molecular to the cellular level, CVDs remain one of the leading causes of death in humans. circRNAs mainly function as a “sponge” to absorb microRNAs, which results in the positive control of downstream proteins. They play important regulatory roles in the development of CVDs. This paper reviews current knowledge on the biogenesis, detection and validation, translation, translocation and degradation, and general functions of circRNAs, with a focus on their roles in CVDs.




Similar content being viewed by others
References
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, Hou L, Wang Y, Wang J (2018) A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 8:5855–5869. https://doi.org/10.7150/thno.27285
Karra R, Poss KD (2017) Redirecting cardiac growth mechanisms for therapeutic regeneration. J Clin Invest 127:427–436. https://doi.org/10.1172/JCI89786
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73:3852–3856. https://doi.org/10.1073/pnas.73.11.3852
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. https://doi.org/10.1261/rna.035667.112
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211. https://doi.org/10.1038/nrm.2015.32
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. https://doi.org/10.1038/nature11993
Zang J, Lu D, Xu A (2020) The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res 98:87–97. https://doi.org/10.1002/jnr.24356
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858. https://doi.org/10.1093/nar/gkw027
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I (2017) Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(22–37):e9. https://doi.org/10.1016/j.molcel.2017.02.017
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. https://doi.org/10.1016/j.molcel.2013.08.017
Gao J, Chen X, Shan C, Wang Y, Li P, Shao K (2021) Autophagy in cardiovascular diseases: role of noncoding RNAs. Mol Ther Nucleic Acids 23:101–118. https://doi.org/10.1016/j.omtn.2020.10.039
Dong R, Ma XK, Chen LL, Yang L (2017) Increased complexity of circRNA expression during species evolution. RNA Biol 14:1064–1074. https://doi.org/10.1080/15476286.2016.1269999
Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. https://doi.org/10.1126/science.aam8526
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. https://doi.org/10.1016/j.molcel.2015.03.027
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. https://doi.org/10.1038/nbt.2890
Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, Dudekula DB, Abdelmohsen K, Gorospe M (2017) High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 45:e116. https://doi.org/10.1093/nar/gkx297
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z (2017) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27:626–641. https://doi.org/10.1038/cr.2017.31
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, Wang F, Ma NF, Guan X, Yun JP, Wang FW, Xu RH, Dan X (2019) N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 10:4695. https://doi.org/10.1038/s41467-019-12651-2
Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK (2019) Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell 74(494–507):e8. https://doi.org/10.1016/j.molcel.2019.02.034
Jia R, Xiao MS, Li Z, Shan G, Huang C (2019) Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov 5:45. https://doi.org/10.1038/s41421-019-0113-y
Huang C, Liang D, Tatomer DC, Wilusz JE (2018) A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32:639–644. https://doi.org/10.1101/gad.314856.118
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M, Li L, Guo H (2019) Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer 18:111. https://doi.org/10.1186/s12943-019-1040-0
Kristensen LS, Hansen TB, Veno MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37:555–565. https://doi.org/10.1038/onc.2017.361
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. https://doi.org/10.1038/ncomms11215
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, Jia L, Zhou L, Li W, Hoffman AR, Hu JF, Cui J (2018) A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol 19:218. https://doi.org/10.1186/s13059-018-1594-y
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77:2339–2350. https://doi.org/10.1158/0008-5472.CAN-16-1883
Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N, Blandino G (2017) The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol 18:237. https://doi.org/10.1186/s13059-017-1368-y
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–1030. https://doi.org/10.1016/0092-8674(93)90279-y
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, Kuo HC (2017) The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 8:1149. https://doi.org/10.1038/s41467-017-01216-w
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL (2017) Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67:214-227.e7. https://doi.org/10.1016/j.molcel.2017.05.023
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR (2015) 5’ UTR m(6)A promotes cap-independent translation. Cell 163:999–1010. https://doi.org/10.1016/j.cell.2015.10.012
van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, Kanda M, Worth CL, Schafer S, Calviello L, Merriott R, Patone G, Hummel O, Wyler E, Obermayer B, Mucke MB, Lindberg EL, Trnka F, Memczak S, Schilling M, Felkin LE, Barton PJR, Quaife NM, Vanezis K, Diecke S, Mukai M, Mah N, Oh SJ, Kurtz A, Schramm C, Schwinge D, Sebode M, Harakalova M, Asselbergs FW, Vink A, de Weger RA, Viswanathan S, Widjaja AA, Gartner-Rommel A, Milting H, Dos Remedios C, Knosalla C, Mertins P, Landthaler M, Vingron M, Linke WA, Seidman JG, Seidman CE, Rajewsky N, Ohler U, Cook SA, Hubner N (2019) The translational landscape of the human heart. Cell 178(242–260):e29. https://doi.org/10.1016/j.cell.2019.05.010
Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S (2016) Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 98:103–107. https://doi.org/10.1016/j.yjmcc.2016.07.007
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H, Shi P, Li J, Gao L, Tian X (2020) Circular RNA profiling reveals exosomal circ_0006156 as a novel biomarker in papillary thyroid cancer. Mol Ther Nucleic Acids 19:1134–1144. https://doi.org/10.1016/j.omtn.2019.12.025
Wang Y, Zhao R, Liu W, Wang Z, Rong J, Long X, Liu Z, Ge J, Shi B (2019) Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev 2019:7954657. https://doi.org/10.1155/2019/7954657
Mendis S, Davis S, Norrving B (2015) Organizational update: the world health organization global status report on noncommunicable diseases 2014; one more landmark step in the combat against stroke and vascular disease. Stroke 46:e121–e122. https://doi.org/10.1161/STROKEAHA.115.008097
Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C (2016) Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteom Bioinform 14:216–223. https://doi.org/10.1016/j.gpb.2016.02.003
Aaronson KD, Sackner-Bernstein J (2006) Risk of death associated with nesiritide in patients with acutely decompensated heart failure. JAMA 296:1465–1466. https://doi.org/10.1001/jama.296.12.1465
Tham YK, Bernardo BC, Ooi JY, Weeks KL, McMullen JR (2015) Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol 89:1401–1438. https://doi.org/10.1007/s00204-015-1477-x
Nakamura M, Sadoshima J (2018) Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol 15:387–407. https://doi.org/10.1038/s41569-018-0007-y
Jahn C, Bar C, Thum T (2019) CircSlc8a1, breaking a vicious circle in cardiac hypertrophy. Cardiovasc Res 115:1946–1947. https://doi.org/10.1093/cvr/cvz147
Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL, Annadoray L, Sian S, Ackers-Johnson MA, Foo RS (2019) Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res 115:1998–2007. https://doi.org/10.1093/cvr/cvz130
Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, Yuan SJ, Zeng N, Yang ZZ, Yang H, Wang XP, Duan JZ, Wang S, Luo JF, Wu SL, Shan ZX (2020) Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res 116:1323–1334. https://doi.org/10.1093/cvr/cvz215
Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611. https://doi.org/10.1093/eurheartj/ehv713
Xu X, Wang J, Wang X (2020) Silencing of circHIPK3 inhibits pressure overload-induced cardiac hypertrophy and dysfunction by sponging miR-185-3p. Drug Des Dev Ther 14:5699–5710. https://doi.org/10.2147/DDDT.S245199
Holdt LM, Teupser D (2012) Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol 32:196–206. https://doi.org/10.1161/ATVBAHA.111.232678
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6:e1001233. https://doi.org/10.1371/journal.pgen.1001233
Zhang S, Song G, Yuan J, Qiao S, Xu S, Si Z, Yang Y, Xu X, Wang A (2020) Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFbetaR2/phosph-SMAD3 axis. J Biomed Sci 27:11. https://doi.org/10.1186/s12929-019-0595-9
Wei MY, Lv RR, Teng Z (2020) Circular RNA circHIPK3 as a novel circRNA regulator of autophagy and endothelial cell dysfunction in atherosclerosis. Eur Rev Med Pharmacol Sci 24:12849–12858. https://doi.org/10.26355/eurrev_202012_24187
Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, Santel A (2005) The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci 118:3049–3059. https://doi.org/10.1242/jcs.02415
Huang S, Li X, Zheng H, Si X, Li B, Wei G, Li C, Chen Y, Chen Y, Liao W, Liao Y, Bin J (2019) Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139:2857–2876. https://doi.org/10.1161/CIRCULATIONAHA.118.038361
Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, Tang Y, Mallaredy V, Ibetti J, Grisanti L, Schumacher SM, Gao E, Rajan S, Wilusz JE, Goukassian D, Houser SR, Koch WJ, Kishore R (2019) Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 10:4317. https://doi.org/10.1038/s41467-019-11777-7
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, Shen G, Wang F (2019) Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol 292:188–196. https://doi.org/10.1016/j.ijcard.2019.04.006
Deng Y-Y, Zhang W, She J, Zhang L, Chen T, Zhou J, Yuan Z (2016) GW27-e1167 circular RNA related to PPARγ function as ceRNA of microRNA in human acute myocardial infarction. J Am Coll Cardiol 68:C51–C52. https://doi.org/10.1016/j.jacc.2016.07.189
Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, Xu J, Chen S, Liu S (2019) Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol 130:10–22. https://doi.org/10.1016/j.yjmcc.2019.03.007
Gupta SK, Garg A, Bar C, Chatterjee S, Foinquinos A, Milting H, Streckfuss-Bomeke K, Fiedler J, Thum T (2018) Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res 122:246–254. https://doi.org/10.1161/CIRCRESAHA.117.311335
Torrealba N, Aranguiz P, Alonso C, Rothermel BA, Lavandero S (2017) Mitochondria in structural and functional cardiac remodeling. Adv Exp Med Biol 982:277–306. https://doi.org/10.1007/978-3-319-55330-6_15
Salgado-Somoza A, Zhang L, Vausort M, Devaux Y (2017) The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 17:33–36. https://doi.org/10.1016/j.ijcha.2017.11.001
Zou M, Huang C, Li X, He X, Chen Y, Liao W, Liao Y, Sun J, Liu Z, Zhong L, Bin J (2017) Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget 8:81825–81837. https://doi.org/10.18632/oncotarget.18998
Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP (2016) The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE 11:e0151753. https://doi.org/10.1371/journal.pone.0151753
Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, Zhang RC, Liu CY, Dong YH, Wang M, Qian LL, Ponnusamy M, Zhang YH, Zhang J, Wang K (2019) The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ 26:1299–1315. https://doi.org/10.1038/s41418-018-0206-4
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF (2017) Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 24:1111–1120. https://doi.org/10.1038/cdd.2017.61
Zhou B, Yu JW (2017) A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun 487:769–775. https://doi.org/10.1016/j.bbrc.2017.04.044
Kurz DJ, Decary S, Hong Y, Erusalimsky JD (2000) Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci 113(Pt 20):3613–3622
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412. https://doi.org/10.1093/eurheartj/ehw001
Nihei T, Takahashi J, Hao K, Kikuchi Y, Odaka Y, Tsuburaya R, Nishimiya K, Matsumoto Y, Ito K, Miyata S, Sakata Y, Shimokawa H (2018) Prognostic impacts of Rho-kinase activity in circulating leucocytes in patients with vasospastic angina. Eur Heart J 39:952–959. https://doi.org/10.1093/eurheartj/ehx657
Enroth S, Johansson A, Enroth SB, Gyllensten U (2014) Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun 5:4684. https://doi.org/10.1038/ncomms5684
Shen L, Hu Y, Lou J, Yin S, Wang W, Wang Y, Xia Y, Wu W (2019) CircRNA0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR107. Mol Med Rep 19:3923–3932. https://doi.org/10.3892/mmr.2019.10011
Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, Wang L (2019) Upregulation of circular RNA CircNFIB Attenuates cardiac fibrosis by sponging miR-433. Front Genet 10:564. https://doi.org/10.3389/fgene.2019.00564
Siede D, Rapti K, Gorska AA, Katus HA, Altmuller J, Boeckel JN, Meder B, Maack C, Volkers M, Muller OJ, Backs J, Dieterich C (2017) Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol 109:48–56. https://doi.org/10.1016/j.yjmcc.2017.06.015
Funding
This work was supported by the National Natural Science Foundation of China (82070314), (81770275), the Key Research and Development Project of Shandong Province (2017GSF18127), and the Taishan Scholar Program of Shandong Province;
Author information
Authors and Affiliations
Contributions
WK put forward the theme of circRNA and CVDs. JJ and LC developed the scope of the review. SY, WT and CX carried out the PubMed searches. JJ summarized the relevant papers, accomplished the figures and wrote the manuscript. WK compiled and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ju, J., Song, Yn., Chen, Xz. et al. circRNA is a potential target for cardiovascular diseases treatment. Mol Cell Biochem 477, 417–430 (2022). https://doi.org/10.1007/s11010-021-04286-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04286-z