Abstract
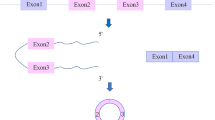
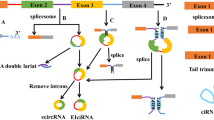
Circular RNA (circRNA) has a closed-loop structure, and its 3’ and 5’ ends are directly covalently connected by reverse splicing, which is more stable than linear RNA. CircRNAs usually possess microRNA (miRNA) binding sites, which can bind miRNAs and inhibit miRNA function. Many studies have shown that circRNAs are involved in the processes of cell senescence, proliferation and apoptosis and a series of signalling pathways, playing an important role in the prevention and treatment of diseases. CircRNAs are potential biological diagnostic markers and therapeutic targets for cardiovascular diseases (CVDs). To identify biomarkers and potential effective therapeutic targets without toxicity for heart disease, we summarize the biogenesis, biology, characterization and functions of circRNAs in CVDs, hoping that this information will shed new light on the prevention and treatment of CVDs.



Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- circRNA:
-
Circular RNA
- miRNA:
-
MicroRNA
- CVD:
-
Cardiovascular disease
- ncRNA:
-
Noncoding RNA
- RBP:
-
RNA binding protein
- RNAPol:
-
RNA polymerase
- ecRNA:
-
Exon circRNA
- ciRNA:
-
Intronic circRNA
- elciRNA:
-
Exon–intron circRNA
- MRE:
-
MiRNA response element
- m6A:
-
N6-methyladenosine
- PKR:
-
Double-stranded RNA-dependent protein kinase
- VSMC:
-
Vascular smooth muscle cell
- EMT:
-
Epithelial-mesenchymal transition
- PES 1:
-
Pescadillo homologue 1
- RCA:
-
Rolling circle amplification
- IRES:
-
Internal ribosome entry site
- MFACR:
-
Mitochondrial fission and apoptosis-related circRNA
- EIF4A3:
-
Eukaryotic translation initiation factor 4A3
- MICRA:
-
Myocardial infarction-related circRNA
- I/R:
-
Ischaemia/reperfusion
- RISK:
-
Reperfusion injury saving kinase
- HRCR:
-
Heart-related circRNA
- SRF:
-
Serum responsive factor
- CTGF:
-
Connective tissue growth factor
- Adrb1:
-
Adrenergic receptor β1
- ADCY6:
-
Adenylate cyclase 6
- TGF:
-
Transforming growth factor
- AF:
-
Atrial fibrillation
- EH:
-
Essential hypertension
- SH:
-
Secondary hypertension
- AAA:
-
Abdominal aortic aneurysm
- BNP:
-
B-type natriuretic peptide
- ANP:
-
Atrial natriuretic peptide
- CRP:
-
C-reactive protein
- IL-6:
-
Interleukin-6
- qPCR:
-
Quantitative polymerase chain reaction
- FISH:
-
Fluorescence in situ hybridization
References
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular rnas are a large class of animal rnas with regulatory potency. Nature. 2013;495(7441):333–8.
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular rna molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–6.
Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of rna in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene sry in adult mouse testis. Cell. 1993;73(5):1019–30.
Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular rna molecules. FASEB J. 1993;7(1):155–60.
Chen X, Han P, Zhou T, Guo X, Song X, Li Y. Circrnadb: a comprehensive database for human circular rnas with protein-coding annotations. Sci Rep. 2016;6:34985.
Abdelmohsen K, Panda A, Munk R, Grammatikakis I, Dudekula D, De S, et al. Identification of hur target circular rnas uncovers suppression of pabpn1 translation by circpabpn1. RNA Biol. 2017;14(3):361–9.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. Circrna biogenesis competes with pre-mrna splicing. Mol Cell. 2014;56(1):55–66.
Ying C, Guohai Y, Ye Z, et al. High glucose-induced circhipk3 downregulation mediates endothelial cell injury. Biochem Biophys Res Commun. 2018;507(1–4):362–8.
Lu D, Thum T. Rna-based diagnostic and therapeutic strategies for cardiovascular disease. Nat Rev Cardiol. 2019;16(11):661–74.
Zhang TR, Huang WQ. Angiogenic circular rnas: a new landscape in cardiovascular diseases. Microvasc Res. 2020;129:103983.
Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular rnas. Wiley Interdiscip Rev RNA. 2015;6(5):563–79.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular rnas regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64.
Kelly SGC, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427(15):2414–7.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding rnas. Mol Cell. 2013;51(6):792–806.
Vo J, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular rna in cancer. Cell. 2019;176(4):869-81.e13.
Suzuki H, Tsukahara T. A view of pre-mrna splicing from rnase r resistant rnas. Int J Mol Sci. 2014;15(6):9331–42.
Du W, Yang W, Chen Y, Wu Z, Foster F, Yang Z, et al. Foxo3 circular rna promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–12.
Cai J, Chen Z, Wang J, Wang J, Chen X, Liang L, et al. Circhectd1 facilitates glutaminolysis to promote gastric cancer progression by targeting mir-1256 and activating β-catenin/c-myc signaling. Cell Death Dis. 2019;10(8):576.
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular rnas in cancer. Cancer Lett. 2018;414:301–9.
Wen G, Zhou T, Gu W. The potential of using blood circular rna as liquid biopsy biomarker for human diseases. Protein Cell. 2020.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular rna-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–17.
Hansen T, Wiklund E, Bramsen J, Villadsen S, Statham A, Clark S, et al. Mirna-dependent gene silencing involving ago2-mediated cleavage of a circular antisense rna. EMBO J. 2011;30(21):4414–22.
Park O, Ha H, Lee Y, Boo S, Kwon D, Song H, et al. Endoribonucleolytic cleavage of ma-containing rnas by rnase p/mrp complex. Mol Cell. 2019;74(3):494-507.e8.
Fischer J, Busa V, Shao Y, Leung A. Structure-mediated rna decay by upf1 and g3bp1. Mol Cell. 2020;78(1):70-84.e6.
Jia R, Xiao M, Li Z, Shan G, Huang C. Defining an evolutionarily conserved role of gw182 in circular rna degradation. Cell Discov. 2019;5:45.
Liu C, Li X, Nan F, Jiang S, Gao X, Guo S, et al. Structure and degradation of circular rnas regulate pkr activation in innate immunity. Cell. 2019;177(4):865-80.e21.
Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen L-L, et al. The output of protein-coding genes shifts to circular rnas when the pre-mrna processing machinery is limiting. Mol Cell. 2017;68(5):940-54.e3.
Beermann J, Piccoli M, Viereck J, Thum T. Non-coding rnas in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–325.
Ebert MS, Neilson JR, Sharp PA. Microrna sponges: competitive inhibitors of small rnas in mammalian cells. Nat Methods. 2007;4(9):721–6.
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mrnas regulates tumour biology. Nature. 2010;465(7301):1033–8.
Zhao F, Chen T, Jiang N. Cdr1as/mir-7/ckap4 axis contributes to the pathogenesis of abdominal aortic aneurysm by regulating the proliferation and apoptosis of primary vascular smooth muscle cells. Exp Ther Med. 2020;19(6):3760–6.
Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular rna locus causes mirna deregulation and affects brain function. Science. 2017;357(6357):eaam8526.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural rna circles function as efficient microrna sponges. Nature. 2013;495(7441):384–8.
Quan H, Chen Q, Wang K, Wang Q, Lu M, Zhang Y, et al. Exendin-4 reversed the pc12 cell damage induced by circrna cdr1as/mir-671/gsk3β signaling pathway. J Mol Neurosci. 2020;71(4):778–89.
Dang R, Liu F, Li Y. Circular rna hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the mir-186/hif-1α axis. Biochem Biophys Res Commun. 2017;490(2):104–10.
Zhuang J, Li T, Hu X, Ning M, Gao W, Lang Y, et al. Circ_chfr expedites cell growth, migration and inflammation in ox-ldl-treated human vascular smooth muscle cells via the mir-214-3p/wnt3/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2020;24(6):3282–92.
Bressin A, Schulte-Sasse R, Figini D, Urdaneta EC, Beckmann BM, Marsico A. Tripepsvm: de novo prediction of rna-binding proteins based on short amino acid motifs. Nucleic Acids Res. 2019;47(9):4406–17.
Zang J, Lu D, Xu A. The interaction of circrnas and rna binding proteins: an important part of circrna maintenance and function. J Neurosci Res. 2020;98(1):87–97.
Ma S, Kong S, Wang F, Ju S. Circrnas: biogenesis, functions, and role in drug-resistant tumours. Mol Cancer. 2020;19(1):119.
Zeng Y, Du W, Wu Y, Yang Z, Awan F, Li X, et al. A circular rna binds to and activates akt phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–55.
Conn S, Pillman K, Toubia J, Conn V, Salmanidis M, Phillips C, et al. The rna binding protein quaking regulates formation of circrnas. Cell. 2015;160(6):1125–34.
Holdt L, Stahringer A, Sass K, Pichler G, Kulak N, Wilfert W, et al. Circular non-coding rna anril modulates ribosomal rna maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular rna in living human cells. Sci Rep. 2015;5:16435.
Santer L, Bär C, Thum T. Circular rnas: a novel class of functional rna molecules with a therapeutic perspective. Mol Ther. 2019;27(8):1350–63.
Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular rnas in human cancer. Mol Cancer. 2020;19(1):30.
Pamudurti N, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circrnas. Mol Cell. 2017;66(1):9-21.e7.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-znf609 is a circular rna that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22-37.e9.
van Heesch S, Witte F, Schneider-Lunitz V, Schulz J, Adami E, Faber A, et al. The translational landscape of the human heart. Cell. 2019;178(1):242-60.e29.
Liu J, Liu T, Wang X, He A. Circles reshaping the rna world: from waste to treasure. Mol Cancer. 2017;16(1):58.
McAloon C, Boylan L, Hamborg T, Stallard N, Osman F, Lim P, et al. The changing face of cardiovascular disease 2000–2012: an analysis of the world health organisation global health estimates data. Int J Cardiol. 2016;224:256–64.
Members WG, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, et al. Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation. 2016;133(4):e38.
Geng HH, Rui L, Su YM, Jie X, Min P, Cai XX, et al. The circular rna cdr1as promotes myocardial infarction by mediating the regulation of mir-7a on its target genes expression. Plos One. 2016;11(3):e0151753.
Wang K, Gan T-Y, Li N, Liu C-Y, Zhou L-Y, Gao J-N, et al. Circular rna mediates cardiomyocyte death via mirna-dependent upregulation of mtp18 expression. Cell Death Differ. 2017;24(6):1111–20.
Lidong Cai, Baozhen Qi, Xiaoyu Wu, et al. Circular rna ttc3 regulates cardiac function after myocardial infarction by sponging mir-15b. J Mol Cell Cardiol. 2019;130:10–22.
Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circrna nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139(25):2857–76.
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting mir-133a-3p. Theranostics. 2018;8(21):5855–69.
Zhou L-Y, Zhai M, Huang Y, Sheng Xu, An T, Wang Y-H, et al. The circular rna acr attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the pink1/fam65b pathway. Cell Death Differ. 2018;26(7):1299–315.
Li Y, Ren S, Xia J, Wei Y, Xi Y. Eif4a3-induced circ-bnip3 aggravated hypoxia-induced injury of h9c2 cells by targeting mir-27a-3p/bnip3. Mol Ther Nucleic Acids. 2020;19:533–45.
Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular rna micra for risk stratification after myocardial infarction. International journal of cardiology. Heart Vasc. 2017;17:33–6.
Bai X, Niu R, Liu J, Pan X, Wang F, Yang W, et al. Roles of noncoding rnas in the initiation and progression of myocardial ischemia-reperfusion injury. Epigenomics. 2021;13(9):715–43.
Song Y, Zhao L, Wang B, Sun J, Hu J, Zhu X, et al. The circular rna tlk1 exacerbates myocardial ischemia/reperfusion injury via targeting mir-214/ripk1 through tnf signaling pathway. Free Radical Biol Med. 2020;155:69–80.
Chen L, Luo W, Zhang W, Chu H, Wang J, Dai X, et al. Circdlpag4/hectd1 mediates ischaemia/reperfusion injury in endothelial cells via er stress. RNA Biol. 2020;17(2):240–53.
Xiao Y, Oumarou DB, Wang S, Liu Y. Circular rna involved in the protective effect of Malva sylvestris l. On myocardial ischemic/re-perfused injury. Front Pharmacol. 2020;11:520486.
Altesha M, Ni T, Khan A, Liu K, Zheng X. Circular rna in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–600.
Zhang CL, Long TY, Bi SS, Sheikh SA, Li F. Circpan3 ameliorates myocardial ischaemia/reperfusion injury by targeting mir-421/pink1 axis-mediated autophagy suppression. Lab Invest. 2021;101(1):89–103.
Wang K, Long B, Liu F, Wang J, Liu C, Zhao B, et al. A circular rna protects the heart from pathological hypertrophy and heart failure by targeting mir-223. Eur Heart J. 2016;37(33):2602–11.
Lim T, Aliwarga E, Luu T, Li Y, Ng S, Annadoray L, et al. Targeting the highly abundant circular rna circslc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. 2019;115(14):1998–2007.
Zhang Y, Chen B. Silencing circ_0062389 alleviates cardiomyocyte apoptosis in heart failure rats via modulating tgf-β1/smad3 signaling pathway. Gene. 2021;766:145154.
Han J, Zhang L, Hu L, Yu H, Xu F, Yang B, et al. Circular rna-expression profiling reveals a potential role of hsa_circ_0097435 in heart failure via sponging multiple micrornas. Front Genet. 2020;11:212.
Deng Y, Wang J, Xie G, Zeng X, Li H. Circ-hipk3 strengthens the effects of adrenaline in heart failure by mir-17-3p - adcy6 axis. Int J Biol Sci. 2019;15(11):2484–96.
Zaiou M. Circular rnas as potential biomarkers and therapeutic targets for metabolic diseases. Adv Exp Med Biol. 2019;1134:177–91.
Zaiou M. Circular rnas in hypertension: challenges and clinical promise. Hypertens Res. 2019;42(11):1653–63.
Zhang S, Chen X, Li C, Li X, Liu C, Liu B, et al. Identification and characterization of circular rnas as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci. 2017;58(14):6500–9.
Zhou B, Yu J. A novel identified circular rna, circrna_010567, promotes myocardial fibrosis via suppressing mir-141 by targeting tgf-β1. Biochem Biophys Res Commun. 2017;487(4):769–75.
Yang F, Li A, Qin Y, Che H, Wang Y, Lv J, et al. A novel circular rna mediates pyroptosis of diabetic cardiomyopathy by functioning as a competing endogenous rna. Mol Ther Nucleic Acids. 2019;17:636–43.
Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr, Woods TC. Carotid plaque rupture is accompanied by an increase in the ratio of serum circr-284 to mir-221 levels. Circ-Cardiovasc Genet. 2017;10(4):e001720.
Ernst C, Odom DT, Kutter C. The emergence of pirnas against transposon invasion to preserve mammalian genome integrity. Nat Commun. 2017;8(1):1411.
Li C, Ma L, Yu B. Circular rna hsa_circ_0003575 regulates oxldl induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother. 2017;95:1514–9.
Kang S, Sohn E, Lee S. Hydrogen sulfide as a potential alternative for the treatment of myocardial fibrosis. Oxid Med Cell Longev. 2020;2020:4105382.
Tang C, Zhang M, Huang L, Hu Z, Zhu J, Xiao Z, et al. Circrna_000203 enhances the expression of fibrosis-associated genes by derepressing targets of mir-26b-5p, col1a2 and ctgf, in cardiac fibroblasts. Sci Rep. 2017;7:40342.
Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, et al. Upregulation of circular rna circnfib attenuates cardiac fibrosis by sponging mir-433. Front Genet. 2019;10:564.
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circhipk3 prevents angiotensin ii-induced cardiac fibrosis by sponging mir-29b-3p. Int J Cardiol. 2019;292:188–96.
Wang K, Dong Y, Liu J, Qian L, Wang T, Gao X, et al. Effects of redox in regulating and treatment of metabolic and inflammatory cardiovascular diseases. Oxid Med Cell Longev. 2020;2020:5860356.
Costa M, Cortez-Dias N, Gabriel A, de Sousa J, Fiúza M, Gallego J, et al. Circrna-mirna cross-talk in the transition from paroxysmal to permanent atrial fibrillation. Int J Cardiol. 2019;290:134–7.
Jiang S, Guo C, Zhang W, Che W, Zhang J, Zhuang S, et al. The integrative regulatory network of circrna, microrna, and mrna in atrial fibrillation. Front Genet. 2019;10:526.
Hu X, Chen L, Wu S, Xu K, Jiang W, Qin M, et al. Integrative analysis reveals key circular rna in atrial fibrillation. Front Genet. 2019;10:108.
Liu T, Zhang G, Wang Y, Rao M, Zhang Y, Guo A, et al. Identification of circular rna-microrna-messenger rna regulatory network in atrial fibrillation by integrated analysis. Biomed Res Int. 2020;2020:8037273.
Zhang P, Sun J, Li W. Genome-wide profiling reveals atrial fibrillation-related circular rnas in atrial appendages. Gene. 2020;728:144286.
Huang Y, Tang C, Du J, Jin H. Endogenous sulfur dioxide: a new member of gasotransmitter family in the cardiovascular system. Oxid Med Cell Longev. 2016;2016:8961951.
Heagerty A, Heerkens E, Izzard A. Small artery structure and function in hypertension. J Cell Mol Med. 2010;14(5):1037–43.
Yin L, Yao J, Deng G, Wang X, Cai W, Shen J. Identification of candidate lncrnas and circrnas regulating wnt3/β-catenin signaling in essential hypertension. Aging. 2020;12(9):8261–88.
Wu N, Jin L, Cai J. Profiling and bioinformatics analyses reveal differential circular rna expression in hypertensive patients. Clin Exp Hypertens. 2017;39(5):454–9.
Bao X, He X, Zheng S, Sun J, Luo Y, Tan R, et al. Up-regulation of circular rna hsa_circ_0037909 promotes essential hypertension. J Clin Lab Anal. 2019;33(4):e22853.
Liu L, Gu T, Bao X, Zheng S, Zhao J, Zhang L. Microarray profiling of circular rna identifies hsa_circ_0126991 as a potential risk factor for essential hypertension. Cytogenet Genome Res. 2019;157(4):203–12.
Prestes P, Maier M, Woods B, Charchar F. A guide to the short, long and circular rnas in hypertension and cardiovascular disease. Int J Mol Sci. 2020;21(10):3666.
Wang J, Sun H, Zhou Y, Huang K, Que J, Peng Y, et al. Circular rna microarray expression profile in 3,4-benzopyrene/angiotensin ii-induced abdominal aortic aneurysm in mice. J Cell Biochem. 2019;120(6):10484–94.
Wilmink A, Quick C. Epidemiology and potential for prevention of abdominal aortic aneurysm. Br J Surg. 1998;85(2):155–62.
Golledge J, Moxon J, Singh T, Bown M, Mani K, Wanhainen A. Lack of an effective drug therapy for abdominal aortic aneurysm. J Intern Med. 2020;288(1):6–22.
Yang R, Wang Z, Meng G, Hua L. Circular rna ccdc66 facilitates abdominal aortic aneurysm through the overexpression of ccdc66. Cell Biochem Funct. 2020;38(7):830–8.
Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y, et al. Circcbfb-mediated mir-28–5p facilitates abdominal aortic aneurysm via lypd3 and gria4. Life Sci. 2020;253:117533.
Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, et al. Cyclic rna hsa-circ-000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep. 2015;12(5):6656–62.
Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular rna wdr77 target fgf-2 to regulate vascular smooth muscle cells proliferation and migration by sponging mir-124. Biochem Biophys Res Commun. 2017;494:126–32.
Mao Y, Wang J, Guo X, Bi Y, Wang C. Circ-satb2 upregulates stim1 expression and regulates vascular smooth muscle cell proliferation and differentiation through mir-939. Biochem Biophys Res Commun. 2018;505(1):119–25.
Zhou M, Shi Z, Cai L, Li X, Ding Y, Xie T, et al. Circular rna expression profile and its potential regulative role in human abdominal aortic aneurysm. BMC Cardiovasc Disord. 2020;20(1):70.
Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular rna micra for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. 2017;17:33–6.
Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Devaux Y. Myocardial infarction-associated circular rna predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68(11):1247–8.
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, et al. Peripheral blood circular rna hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918.
Yang Y, Yu T, Jiang S, Zhang Y, Li M, Tang N, et al. Mirnas as potential therapeutic targets and diagnostic biomarkers for cardiovascular disease with a particular focus on wo2010091204. Expert Opin Ther Pat. 2017;27(9):1021–9.
Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, et al. A potential risk factor of essential hypertension in case-control study: circular rna hsa_circ_0037911. Biochem Biophys Res Commun. 2018;498(4):789–94.
Garikipati V, Verma S, Cheng Z, Liang D, Truongcao M, Cimini M, et al. Circular rna circfndc3b modulates cardiac repair after myocardial infarction via fus/vegf-a axis. Nat Commun. 2019;10(1):4317.
Shen L, Hu Y, Lou J, Yin S, Wang W, Wang Y, et al. Circrna-0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting mir-107. Mol Med Rep. 2019;19(5):3923–32.
Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular rna circchfr facilitates the proliferation and migration of vascular smooth muscle via mir-370/foxo1/cyclin d1 pathway. Mol Ther Nucleic Acids. 2019;16:434–41.
Zhang Y, Chen Y, Yao H, Lie Z, Chen G, Tan H, et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ. 2019;9(4):2045894019888416.
Zheng S, Gu T, Bao X, Sun J, Zhao J, Zhang T, et al. Circular rna hsa_circ_0014243 may serve as a diagnostic biomarker for essential hypertension. Exp Ther Med. 2019;17(3):1728–36.
Liu Y, Yang Y, Wang Z, Fu X, Chu X, Li Y, et al. Insights into the regulatory role of circrna in angiogenesis and clinical implications. Atherosclerosis. 2020;298:14–26.
Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular rnas in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:5135781.
Odqvist M, Andersson P, Tygesen H, Eggers K, Holzmann M. High-sensitivity troponins and outcomes after myocardial infarction. J Am Coll Cardiol. 2018;71(23):2616–24.
McDonald K, Troughton R, Dahlström U, Dargie H, Krum H, van der Meer P, et al. Daily home bnp monitoring in heart failure for prediction of impending clinical deterioration: results from the home hf study. Eur J Heart Fail. 2018;20(3):474–80.
Gaggin H, Januzzi J. Biomarkers and diagnostics in heart failure. Biochem Biophys Acta. 2013;1832(12):2442–50.
Marciniak A, Nawrocka Rutkowska J, Brodowska A, Wiśniewska B, Starczewski A. Cardiovascular system diseases in patients with polycystic ovary syndrome - the role of inflammation process in this pathology and possibility of early diagnosis and prevention. Ann Agric Environ Med. 2016;23(4):537–41.
Olson J. D-dimer: an overview of hemostasis and fibrinolysis, assays, and clinical applications. Adv Clin Chem. 2015;69:1–46.
Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circrnas in gastric cancer. Mol Cancer. 2019;18(1):136.
Boeckel J, Jaé N, Heumüller A, Chen W, Boon R, Stellos K, et al. Identification and characterization of hypoxia-regulated endothelial circular rna. Circ Res. 2015;117(10):884–90.
Yang J, Cheng M, Gu B, Wang J, Yan S, Xu D. Circrna_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via mir-6089/akt1/nf-κb axis. Cell Death Dis. 2020;11(10):833.
Fan CY, Lei XJ, Fang ZQ, Jiang QH, Wu FX. Circr2disease: a manually curated database for experimentally supported circular rnas associated with various diseases. Database (Oxford). 2018;2018:bay044.
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular rna promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2018;24(9):1609–20.
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular rnas in the human and mouse genomes. Brief Bioinform. 2017;18(6):984–92.
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. Exorbase: a database of circrna, lncrna and mrna in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–12.
Tang Z, Li ZX, Zhao J, Qian F, Feng C, Li Y, et al. Trcirc: a resource for transcriptional regulation information of circrnas. Brief Bioinform. 2019;20(6):2327–33.
Meng XW, Hu DH, Zhang PJ, Chen Q, Chen M. Circfunbase: a database for functional circular rnas. Database (Oxford). 2019;2019:6.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. Circinteractome: a web tool for exploring circular rnas and their interacting proteins and micrornas. RNA Biol. 2016;13(1):34–42.
Zhao S, Li S, Liu W, Wang Y, Li X, Zhu S, et al. Circular rna signature in lung adenocarcinoma: a mioncocirc database-based study and literature review. Front Oncol. 2020;10:523342.
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2traits: a comprehensive database for circular rna potentially associated with disease and traits. Front Genet. 2013;4:283.
Wu S, Liu H, Huang P, Chang I, Lee C, Yang C, et al. Circlncrnanet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding rnas. GigaScience. 2018;7(1):1–10.
Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circrna with standard nomenclature. RNA Biol. 2019;16(7):899–905.
Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, et al. Circnet: a database of circular rnas derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–15.
Wu W, Ji P, Zhao F. Circatlas: An integrated resource of one million highly accurate circular rnas from 1070 vertebrate transcriptomes. Genome Biol. 2020;21(1):101.
Zheng L, Li J, Wu J, Sun W, Liu S, Wang Z, et al. Deepbase v2.0: identification, expression, evolution and function of small rnas, lncrnas and circular rnas from deep-sequencing data. Nucleic Acids Res. 2016;44(D1):D196-202.
Zhong S, Wang J, Zhang Q, Xu H, Feng J. Circprimer: a software for annotating circrnas and determining the specificity of circrna primers. BMC Bioinformatics. 2018;19(1):292.
Dong R, Ma X, Li G, Yang L. Circpedia v2: an updated database for comprehensive circular rna annotation and expression comparison. Genomics Proteomics Bioinformatics. 2018;16(4):226–33.
Acknowledgements
We thank Professor Kun Wang, the head of the Heart Development Center, and Professor Peifeng Li, the head of translational research, who provided substantial scientific support to this work.
Funding
This work was supported by the National Natural Science Foundation of China (81870236, 82070313, 81770275), Taishan Scholar Programme of Shandong Province, Major Research Programme of the National Natural Science Foundation of China (No. 91849209) and Qingdao Scientific Programme (No. 18–6-1–63-nsh).
Author information
Authors and Affiliations
Contributions
Lu-Yu Zhou, Kai Wang, Xiang-Qian Gao and Tao Wang provided direction and guidance throughout the preparation of this manuscript. Kai Wang drafted the manuscript. Lu-Yu Zhou reviewed and made significant revisions to the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, K., Gao, XQ., Wang, T. et al. The Function and Therapeutic Potential of Circular RNA in Cardiovascular Diseases. Cardiovasc Drugs Ther 37, 181–198 (2023). https://doi.org/10.1007/s10557-021-07228-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10557-021-07228-5