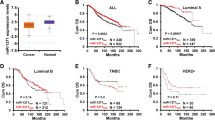
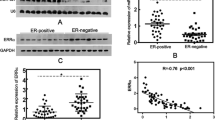
Abstract
TWIST1 (Twist) is a basic helix-loop-helix transcription factor that is overexpressed in many cancers and promotes tumor cell invasion, metastasis, and recurrence. In this study, we demonstrate that Twist upregulates expression of microRNA 22 (miR-22) which, in turn, downregulates estrogen receptor alpha (ER) expression in breast cancer. Initial analysis of miR-22 and Twist expression in a panel of breast cancer cell lines showed a direct correlation between Twist and miR-22 levels with miR-22 being highly expressed in ER negative cell lines. Overexpressing Twist caused increased miR-22 levels while downregulating it led to decreased miR-22 expression. To characterize the upstream promoter region of miR-22, we utilized rapid amplification of cDNA ends and identified the transcription start site and the putative promoter region of miR-22. Mechanistically, we determined that Twist, in combination with HDAC1 and DNMT3B, transcriptionally upregulates miR-22 expression by binding to E-boxes in the proximal miR-22 promoter. We also established that miR-22 causes an increase in growth in 3D but not 2D cultures. Importantly, we observed a direct correlation between increased breast cancer grade and Twist and miR-22 expression. We also identified two potential miR-22 binding sites in the 3′-UTR region of ER and confirmed by promoter assays that miR-22 regulates ER expression by binding to both target sites. These results reveal a novel pathway of ER suppression by Twist through miR-22 activation that could potentially promote the ER negative phenotype in breast cancers.






Similar content being viewed by others
Availability of data and material
Freely available.
References
Hamamori Y, Wu HY, Sartorelli V, Kedes L (1997) The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol Cell Biol 17(11):6563–6573. https://doi.org/10.1128/mcb.17.11.6563
Jan YN, Jan LY (1993) HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75(5):827–830. https://doi.org/10.1016/0092-8674(93)90525-u
Spicer DB, Rhee J, Cheung WL, Lassar AB (1996) Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272(5267):1476–1480. https://doi.org/10.1126/science.272.5267.1476
Castanon I, Baylies MK (2002) A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287(1–2):11–22. https://doi.org/10.1016/s0378-1119(01)00893-9
Castanon I, Von Stetina S, Kass J, Baylies MK (2001) Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128(16):3145–3159
Oshima A, Tanabe H, Yan T, Lowe GN, Glackin CA, Kudo A (2002) A novel mechanism for the regulation of osteoblast differentiation: transcription of periostin, a member of the fasciclin I family, is regulated by the bHLH transcription factor, twist. J Cell Biochem 86(4):792–804. https://doi.org/10.1002/jcb.10272
Thisse B, el Messal M, Perrin-Schmitt F (1987) The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res 15(8):3439–3453. https://doi.org/10.1093/nar/15.8.3439
Baylies MK, Bate M (1996) Twist: a myogenic switch in Drosophila. Science 272(5267):1481–1484. https://doi.org/10.1126/science.272.5267.1481
Leptin M (1991) Twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5(9):1568–1576
Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN (1998) The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev 12(3):422–434. https://doi.org/10.1101/gad.12.3.422
Hebrok M, Fuchtbauer A, Fuchtbauer EM (1997) Repression of muscle-specific gene activation by the murine Twist protein. Exp Cell Res 232(2):295–303. https://doi.org/10.1006/excr.1997.3541
Gitelman I (1997) Twist protein in mouse embryogenesis. Dev Biol 189(2):205–214. https://doi.org/10.1006/dbio.1997.8614
Technau U, Scholz CB (2003) Origin and evolution of endoderm and mesoderm. Int J Dev Biol 47(7–8):531–539
Lee MS, Lowe GN, Strong DD, Wergedal JE, Glackin CA (1999) TWIST, a basic helix-loop-helix transcription factor, can regulate the human osteogenic lineage. J Cell Biochem 75(4):566–577. https://doi.org/10.1002/(sici)1097-4644(19991215)75:4<566::aid-jcb3>3.0.co;2-0
Li L, Cserjesi P, Olson EN (1995) Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol 172(1):280–292. https://doi.org/10.1006/dbio.1995.0023
Fuchtbauer EM (1995) Expression of M-twist during postimplantation development of the mouse. Dev Dyn 204(3):316–322. https://doi.org/10.1002/aja.1002040309
Chen ZF, Behringer RR (1995) Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev 9(6):686–699. https://doi.org/10.1101/gad.9.6.686
O’Rourke MP, Tam PP (2002) Twist functions in mouse development. Int J Dev Biol 46(4):401–413
Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ (1999) Twist is a potential oncogene that inhibits apoptosis. Genes Dev 13(17):2207–2217. https://doi.org/10.1101/gad.13.17.2207
Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P Jr, Raman V (2005) HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J Biol Chem 280(3):2294–2299. https://doi.org/10.1074/jbc.M411018200
Mironchik Y, Winnard PT Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, Burger H, Glackin C, Raman V (2005) Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res 65(23):10801–10809. https://doi.org/10.1158/0008-5472.CAN-05-0712
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117(7):927–939. https://doi.org/10.1016/j.cell.2004.06.006
Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF (2002) Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol 161(5):1881–1891. https://doi.org/10.1016/S0002-9440(10)64464-1
Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R (2004) Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res 64(15):5270–5282. https://doi.org/10.1158/0008-5472.CAN-04-0731
van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP (2004) Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis. Cancer Res 64(16):5578–5586. https://doi.org/10.1158/0008-5472.CAN-04-1253
Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X (2005) Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res 65(12):5153–5162. https://doi.org/10.1158/0008-5472.CAN-04-3785
Campbell RJ, Pignatelli M (2002) Molecular histology in the study of solid tumours. Mol Pathol 55(2):80–82. https://doi.org/10.1136/mp.55.2.80
Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2(2):76–83. https://doi.org/10.1038/35000025
Woodhouse EC, Chuaqui RF, Liotta LA (1997) General mechanisms of metastasis. Cancer 80(8 Suppl):1529–1537. https://doi.org/10.1002/(sici)1097-0142(19971015)80:8+<1529::aid-cncr2>3.3.co;2-#
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64(11):3753–3756. https://doi.org/10.1158/0008-5472.CAN-04-0637
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435(7043):839–843. https://doi.org/10.1038/nature03677
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103(7):2257–2261. https://doi.org/10.1073/pnas.0510565103
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101(9):2999–3004. https://doi.org/10.1073/pnas.0307323101
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838. https://doi.org/10.1038/nature03702
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070. https://doi.org/10.1158/0008-5472.CAN-05-1783
Hossain A, Kuo MT, Saunders GF (2006) Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol 26(21):8191–8201. https://doi.org/10.1128/MCB.00242-06
Hurteau GJ, Carlson JA, Spivack SD, Brock GJ (2007) Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res 67(17):7972–7976. https://doi.org/10.1158/0008-5472.CAN-07-1058
Adams BD, Furneaux H, White BA (2007) The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21(5):1132–1147. https://doi.org/10.1210/me.2007-0022
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47(D1):D155–D162. https://doi.org/10.1093/nar/gky1141
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45. https://doi.org/10.1093/nar/29.9.e45
Vesuna F, Lisok A, Kimble B, Domek J, Kato Y, van der Groep P, Artemov D, Kowalski J, Carraway H, van Diest P, Raman V (2012) Twist contributes to hormone resistance in breast cancer by downregulating estrogen receptor-alpha. Oncogene 31(27):3223–3234. https://doi.org/10.1038/onc.2011.483
Church GM, Ephrussi A, Gilbert W, Tonegawa S (1985) Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature 313(6005):798–801. https://doi.org/10.1038/313798a0
Ephrussi A, Church GM, Tonegawa S, Gilbert W (1985) B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science 227(4683):134–140. https://doi.org/10.1126/science.3917574
Pandey DP, Picard D (2009) miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 29(13):3783–3790. https://doi.org/10.1128/MCB.01875-08
Vesuna F, Lisok A, Kimble B, Raman V (2009) Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia 11(12):1318–1328. https://doi.org/10.1593/neo.91084
Vesuna F, van Diest P, Chen JH, Raman V (2008) Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun 367(2):235–241. https://doi.org/10.1016/j.bbrc.2007.11.151
Li J, Zhou BP (2011) Activation of beta-catenin and Akt pathways by twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 11:49. https://doi.org/10.1186/1471-2407-11-49
Liu X, Zhang L, Tong Y, Yu M, Wang M, Dong D, Shao J, Zhang F, Niu R, Zhou Y (2019) MicroRNA-22 inhibits proliferation, invasion and metastasis of breast cancer cells through targeting truncated neurokinin-1 receptor and ERalpha. Life Sci 217:57–69. https://doi.org/10.1016/j.lfs.2018.11.057
Shao Y, Yao Y, Xiao P, Yang X, Zhang D (2019) Serum miR-22 could be a potential biomarker for the prognosis of breast cancer. Clin Lab 65(4). https://doi.org/10.7754/Clin.Lab.2018.180825
Xiang Q, Xiang Z, Dou R, Xiong B (2019) Survival advantage and clinicopathological significance of microRNA-22 in cancers: a meta-analysis. Cancer Manag Res 11:8855–8868. https://doi.org/10.2147/CMAR.S185124
Marchesi F, Regazzo G, Palombi F, Terrenato I, Sacconi A, Spagnuolo M, Donzelli S, Marino M, Ercolani C, Di Benedetto A, Blandino G, Ciliberto G, Mengarelli A, Rizzo MG (2018) Serum miR-22 as potential non-invasive predictor of poor clinical outcome in newly diagnosed, uniformly treated patients with diffuse large B-cell lymphoma: an explorative pilot study. J Exper Clin Can Res CR 37(1):95. https://doi.org/10.1186/s13046-018-0768-5
Kim JB (2005) Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol 15(5):365–377. https://doi.org/10.1016/j.semcancer.2005.05.002
Montani C, Steimberg N, Boniotti J, Biasiotto G, Zanella I, Diafera G, Biunno I, Caimi L, Mazzoleni G, Di Lorenzo D (2014) Fibroblasts maintained in 3 dimensions show a better differentiation state and higher sensitivity to estrogens. Toxicol Appl Pharmacol 280(3):421–433. https://doi.org/10.1016/j.taap.2014.08.021
Vantangoli MM, Madnick SJ, Wilson S, Boekelheide K (2016) Estradiol exposure differentially alters monolayer versus microtissue MCF-7 human breast carcinoma cultures. PLoS One 11(7):e0157997. https://doi.org/10.1371/journal.pone.0157997
Acknowledgements
We would like to acknowledge John Domek, Ashley Irving, and Yehudit Bergman for technical help with this work.
Funding
This work was supported by the Maryland Stem Cell Research Fund [MDTSCRC0072 to F.V., MDTSCRC0064 to V.R.] and the National Institutes of Health [R01CA097226 to V.R.].
Author information
Authors and Affiliations
Contributions
FV performed the investigation, conceptualization, methodology, analysis, and writing - original draft preparation. AL performed investigation. PvD contributed to the supply of breast cancer samples. VR performed supervision, reviewing, and editing.
Corresponding authors
Ethics declarations
Ethical approval
We used anonymous archival leftover pathology material sourced from Utrecht Medical School. No ethical approval is required as use of de-identified leftover material is part of the standard agreement with patients in the hospital.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent for publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vesuna, F., Lisok, A., van Diest, P. et al. Twist activates miR-22 to suppress estrogen receptor alpha in breast cancer. Mol Cell Biochem 476, 2295–2306 (2021). https://doi.org/10.1007/s11010-021-04065-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04065-w