Abstract
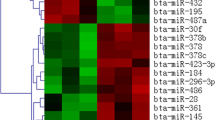
MicroRNAs (miRNAs), a novel class of post-transcriptional gene regulators, have been demonstrated to be involved in several cellular processes regulating the expression of protein-coding genes. To investigate the mechanisms of miRNA-mediated regulation during the process of muscle atrophy, we performed miRNA microarray hybridization between normal differentiated C2C12 cells and dexamethasone (DEX)-treated C2C12 cells. We observed that 11 miRNAs were significantly up-regulated and six miRNAs were down-regulated in the differentiated C2C12 cells after being treated with DEX. Stem–loop real-time RT-PCR confirmed the differential expression of six selected miRNAs (miR-1, miR-147, miR-322, miR-351, and miR-503*, miR-708). miRNA potential target prediction was accomplished using TargetScan, and many target genes related to muscle growth and atrophy have been reported in previous studies. The results of the current study suggested the potential roles of these differentially expressed miRNAs in skeletal muscle atrophy.




Similar content being viewed by others
References
Czerwinski SM, Zak R, Kurowski TT, Falduto MT, Hickson RC (1989) Myosin heavy chain turnover and glucocorticoid deterrence by exercise in muscle. J Appl Physiol 67(6):2311–2315
Adams GR, Caiozzo VJ, Baldwin KM (2003) Skeletal muscle unweighting: spaceflight and ground-based models. J Appl Physiol 95(6):2185–2201
Di Giovanni S, Molon A, Broccolini A, Melcon G, Mirabella M, Hoffman EP (2004) Constitutive activation of MAPK cascade in acute quadriplegic myopathy. Ann Neurol 55(2):195–206
Price DA, Bassendine MF, Norris SM, Golding C, Toms GL, Schmid ML, Morris CM, Burt AD, Donaldson PT (2006) Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut 55(5):715–718
Sever JL, Rakusan TA, Campos JM, O’Donnell RM, Price VA (1996) HIV antibody responses in children of HIV-infected mothers. Pediatr AIDS HIV Infect 7(4):246–253
Miró O, Pedrol E, Cebrián M, Masanés F, Casademont J, Mallolas J, Grau JM (1997) Skeletal muscle studies in patients with HIV-related wasting syndrome. J Neurol Sci 150(2):153–159
Baracos VE (2001) Management of muscle wasting in cancer-associated cachexia: understanding gained from experimental studies. Cancer 92(6 Suppl):1669–1677
Mitch WE, Price SR (2001) Transcription factors and muscle cachexia: is there a therapeutic target? Lancet 357(9258):734–735
Brown M, Hasser EM (1996) Complexity of age-related change in skeletal muscle. J Gerontol A Biol Sci Med Sci 51(2):B2117–B2123
Tawa NE Jr, Odessey R, Goldberg AL (1997) Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest 100:197–203
Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18:39–51
Murton AJ, Constantin D, Greenhaf PL (2008) The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochim Biophys Acta 1782(12):730–743
Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ (2004) The IGF-1/PI3 K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14(3):395–403
Pallafacehina G, Calabria E, Serrano AL (2002) A protein kinase B-dependent andrapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci USA 14(2):432–438
Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ (2001) Mediation of IGF-1-induced skeletal myotube hypertrophy by PI3 K/Akt/mTOR and PI3 K/Akt/GSK3 pathways. Nat Cell Biol 3(11):1009–1013
Cai D, Frantz JD, Tawa NE Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE (2004) IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119(2):285–298
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Kim VN (2004) MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol 14:156–159
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21(17):4663–4670
Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94(6):776–780
Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38(2):228–233
Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND et al (2007) MicroRNA-133 controls cardiac hypertrophy. Nat Med 13(5):613–618
Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M (2007) MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100(3):416–424
Shen H, Zhao SH, Cao JH, Li XY, Fan B (2011) Porcine MuRF2 and MuRF3: molecular cloning, expression and association analysis with muscle production traits. Mol Biol Rep 38(8):5115–5123
ivak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25(4):402–408
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31(4):e15
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Methods Enzymol 411:134–193
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res 33(20):e179
Xie SS, Huang TH, Shen Y, Li XY, Zhang XX, Zhu MJ, Qin HY, Zhao SH (2010) Identification and characterization of microRNAs from porcine skeletal muscle. Anim Genet 41(2):179–190
Mendell JT (2005) MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle 4:1179–1184
Miska EA (2005) How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15:563–568
Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450
Shivdasani RA (2006) MicroRNAs: regulators of gene expression and cell differentiation. Blood 108:3646–3653
Wiemer EA (2007) The role of microRNAs in cancer: no small matter. Eur J Cancer 43:1529–1544
Tan PK, Downey TJ, Spitznagel EL Jr, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC (2003) Evaluation of gene expression measurements from commercial microarray platforms. NucleicAcids Res 31(19):5676–5684
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD (2007) Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120:1046–1054
Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S et al (2007) Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA 104:17016–17021
Allen DL, Loh AS (2011) Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am J Physiol Cell Physiol 300(1):C124–C137
Sartorelli V, Fulco M (2004) Molecular and cellular determinants of skeletal muscle atrophy and hypertrophy. Sci STKE 2004(244):re11
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294(5543):858–862
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M et al (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129(7):1401–1414
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5(3):R13
Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y et al (2005) MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 14(12):2486–2494
Sarkar S, Dey BK, Dutta A (2010) MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell 21(13):2138–2149
Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN, Liu XY, Zhang YY, Li Y, Lin SG et al (2009) Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun 81(4):597–601
Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ et al (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3(11):1014–1019
Ren H, Yin P, Duan C (2008) IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. J Cell Biol 182(5):979–991
Acknowledgments
This study was supported by the National Outstanding Youth Foundation of NSFC (31025026), the NSFC youth foundation (30900792), the creative team project of the Ministry of Education of China (IRT-0831), and the National Key Basic Research Program of China (2012CB124702).
Conflict of interests
The authors have declared that no competing interest exists.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, H., Liu, T., Fu, L. et al. Identification of microRNAs involved in dexamethasone-induced muscle atrophy. Mol Cell Biochem 381, 105–113 (2013). https://doi.org/10.1007/s11010-013-1692-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-013-1692-9