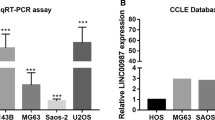
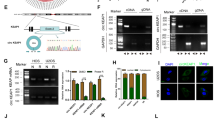
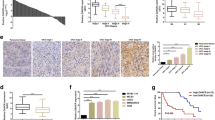
Abstract
Ezrin, one of the ezrin/radixin/moesin (ERM) protein family which act as membrane organizers and linkers between plasma membrane and cytoskeleton, has attracted much attention as a crucial factor for tumor metastasis. Overexpression of ezrin has been correlated with the metastatic potential of several cancers especially for osteosarcoma. Short interfering RNA (siRNA) downregulate gene expression through an enzyme-mediated process named RNA interference (RNAi). RNAi has rapidly come to be recognized as a powerful tool for the study of gene function and a potential target therapy. In the present study, the human osteosarcoma cell line MG63 was cultured. Three siRNAs targeting ezrin mRNA were designed by the multiple computational methods and then were sythesized. These siRNAs were transfected into osteosarcoma cells. Then the expression of ezrin mRNA and protein in osteosarcoma cells was detected. The cellular proliferation and apoptosis was evaluated. C726–U730, C1653–A1661 and G1749–A1771 were selected to be the suitable target sites through the multiple computational methods because of their ideal secondary structures and hybridization thermodynamics. siRNAs against G1749–A1771 downregulated the expression level of ezrin mRNA and protein, inhibit the cellular proliferation and promoted the cellular apoptosis effectively. There is a significant correlation between the multiple computational methods and the efficacy of the corresponding siRNAs. siRNAs targeting ezrin may have therapeutic potential as inhibitors of osteosarcoma metastasis.



Similar content being viewed by others
References
Bielack SS, Kempf-Bielack B, Delling G et al (2002) Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 20:776–790
Damron TA, Ward WG, Stewart A (2007) Osteosarcoma, chondrosarcoma, and Ewing’s sarcoma: National Cancer Data Base report. Clin Orthop Relat Res 459:40–47
Sweetnam R (1982) Osteosarcoma. Brit J Hosp Med 28:16–21
Harris MB, Gieser P, Goorin AM et al (1998) Treatment of metastatic osteosarcoma at diagnosis: a Pediatric Oncology Group Study. J Clin Oncol 16:3641–3648
Meyers PS, Heller G, Healey G et al (1993) Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J Clin Oncol 11:449–453
Khanna C, Khan J, Nguyen P et al (2001) Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res 61:3750–3759
Khanna C, Wan X, Bose S et al (2004) The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med 10:182–186
Krishnan K, Bruce B, Hewitt S et al (2006) Ezrin mediates growth and survival in Ewing’s sarcoma through the AKT/mTOR, but not the MAPK, signaling pathway. Clin Exp Metastasis 23:227–236
Weng WH, Ahlen J, Astrom K et al (2005) Prognostic impact of immunohistochemical expression of ezrin in highly malignant soft tissue sarcomas. Clin Cancer Res 11:6198–6204
Mangeat P, Roy C, Martin M (1999) ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol 9:187–192
Tsukita S, Yonemura S (1997) ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol 9:70–75
Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Ameres SL, Martinez J, Schroeder R (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130:101–112
Bohula EA, Salisbury AJ, Sohail M et al (2003) The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J Biol Chem 278:15991–15997
Kretschmer-Kazemi Far R, Sczakiel G (2003) The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res 31:4417–4424
Schubert S, Grunweller A, Erdmann VA et al (2005) Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol 348:883–893
Vickers TA, Wyatt JR, Freier SM (2000) Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res 28:1340–1347
Zhao JJ, Lemke G (1998) Rules for ribozymes. Mol Cell Neurosci 11:92–97
Overhoff M, Alken M, Far RK et al (2005) Local RNA target structure influences siRNA efficacy: a systematic global analysis. J Mol Biol 348:871–881
Yoshinari K, Miyagishi M, Taira K (2004) Effects on RNAi of the tight structure, sequence, and position of the targeted region. Nucleic Acids Res 32:691–699
Long D, Lee R, Williams P et al (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14:287–294
Zhao Y, Samal E, Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214–220
Denman RB (1993) Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques 15:1090–1095
James W, Cowe E (1997) Computational approaches to the identification of ribozyme target sites. Methods Mol Biol 74:17–26
Sczakiel G, Tabler M (1997) Computer-aided calculation of the local folding potential of target RNA and its use for ribozyme design. Methods Mol Biol 74:11–15
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Chalk AM, Sonnhammer ELL (2002) Computational antisense oligo prediction with a neural network model. Bioinformatics 18:1567–1575
Giddings MC, Shah AA, Freier S et al (2002) Artificial neural network prediction of antisense oligodeoxynucleotide activity. Nucleic Acids Res 30:4295–4304
Saetrom P (2004) Predicting the efficacy of short oligonucleotides in antisense and RNAi experiments with boosted genetic programming. Bioinformatics 20:3055–3063
Thierry AR, Rahman A, Dritschilo A (1993) Overcoming multi drug resistance in human tumor cells using free and liposomally encapsulated antisense oligodeoxynucleotides. Biochem Biophys Res Commun 190:952–960
Jaeger JA, Turner DH, Zuker M et al (1989) Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA 86:7706–7710
Walter AE, Turner DH, Kim J et al (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves prediction of RNA folding. Proc Natl Acad Sci USA 91:9218–9222
Zuker M (1989) On finding all suboptimal foldings of an RNA molecule. Science 244:48–52
Lu ZJ, Mathews DH (2008) Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res 36:640–647
Mathews DH, Burkard ME, Freier SM et al (1999) Predicting oligonucleotide affinity to nucleic acid targets. RNA 5:1458–1469
SantaLucia J, Allawi HT, Seneviratne PA (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35:3555–3562
Sugimoto N, Nakano SI, Katoh M et al (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34:11211–11216
Cairns MJ, Hopkins TM, Witherington C et al (1999) Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol 17:480–486
Lu ZX, Ye M, Yan GR et al (2005) Effect of EBV LMP1 targeted DNAzymes on cell proliferation and apoptosis. Cancer Gene Ther 12:646–654
Pyle AM, Chu VT, Jankowsky E et al (2000) Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol 317:140–146
Mathews DH, Disney MD, Childs JL et al (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA 101:7287–7292
Xia T, SantaLucia J Jr, Burkard ME et al (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick pairs. Biochemistry 37:14719–14735
Torgeir H, Mohammed A, Merete TW et al (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res 30:1757–1766
Beale G, Hollins AJ, Benboubetra M et al (2003) Gene silencing nucleic acids designed by scanning arrays: anti-EGFR activity of siRNA, ribozyme and DNA enzymes targeting a single hybridization-accessible region using the same delivery system. J Drug Target 11:449–456
Petch AK, Sohail M, Hughes MD et al (2003) Messenger RNA expression profiling of genes involved in epidermal growth factor receptor signalling in human cancer cells treated with scanning array-designed antisense oligonucleotides. Biochem Pharmacol 66:819–830
Sohail M, Hochegger H, Klotzbucher A et al (2001) Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res 29:2041–2051
Kurreck J, Bieber B, Jahnel R et al (2002) Comparative study of DNA enzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J Biol Chem 277:7099–7107
Schubert S, Furste JP, Werk D et al (2004) Gaining target access for deoxyribozymes. J Mol Biol 339:355–363
Ho SP, Bao Y, Lesher T et al (1998) Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat Biotechnol 16:59–63
Scherer LJ, Rossi JJ (2003) Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol 21:1457–1465
Laptev AV, Lu Z, Colige A et al (1994) Specific inhibition of expression of a human collagen gene (COL1A1) with modified antisense oligonucleotides. Biochemistry 33:11033–11039
Sczakiel G, Homann M, Rittner K (1993) Computer-aided search for effective antisense RNA target sequences of the human immunodeficiency virus type 1. Antisense Res Dev 3:45–52
Lima WF, Monia BP, Ecker DJ et al (1992) Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry 31:12055–12061
Ding Y, Lawrence CE (2001) Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res 29:1034–1046
Matveeva O, Felden B, Audlin S et al (1997) A rapid in vitro method for obtaining RNA accessibility patterns for complementary DNA probes: correlation with an intracellular pattern and known RNA structures. Nucleic Acids Res 25:5010–5016
Shao Y, Wu S, Chan CY et al (2007) A structural analysis of in vitro catalytic activities of hammerhead ribozymes. BMC Bioinformatics 8:469
Stein CA et al (1999) Keeping the biotechnology of antisense in context. Nat Biotechnol 17:209–212
Acknowledgments
The authors thank Dr. Gang Yao, Dr. Peng Cheng and Dr. JianXue Zeng at the Department of Orthopedic Surgery, Anhui Provincial Hospital Affiliated to Anhui Medical University for helpful discussions, comments, and critical reading of the manuscript.
Conflicts of interest
The authors have no conflicts of interest to disclose.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shang, X., Wang, Y., Zhao, Q. et al. siRNAs target sites selection of ezrin and the influence of RNA interference on ezrin expression and biological characters of osteosarcoma cells. Mol Cell Biochem 364, 363–371 (2012). https://doi.org/10.1007/s11010-012-1238-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-012-1238-6