Abstract
The Z-disc, appearing as a fine dense line forming sarcomere boundaries in striated muscles, when studied in detail reveals crosslinked filament arrays that transmit tension and house myriads of proteins with diverse functions. At the Z-disc the barbed ends of the antiparallel actin filaments from adjoining sarcomeres interdigitate and are crosslinked primarily by layers of α-actinin. The Z-disc is therefore the site of polarity reversal of the actin filaments, as needed to interact with the bipolar myosin filaments in successive sarcomeres. The layers of α-actinin determine the Z-disc width: fast fibres have narrow (~30–50 nm) Z-discs and slow and cardiac fibres have wide (~100 nm) Z-discs. Comprehensive reviews on the roles of the numerous proteins located at the Z-disc in signalling and disease have been published; the aim here is different, namely to review the advances in structural aspects of the Z-disc.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
From a barely visible line in histological sections to a highly dense band in electron micrographs defining the sarcomere boundary in striated muscles, the Z-disc (Z-line, Z-band) on detailed inspection transforms into an intricate landscape of ordered filaments, crosslinks and anchored proteins (Clark et al. 2002; Squire et al. 2005). The filaments are the terminal (“barbed”) ends of the interdigitating, tetragonally arranged, actin filaments from adjoining sarcomeres (Fig. 1). The crosslinks (called Z-links) between the opposite polarity actin filaments are composed mainly of α-actinin molecules (Takahashi and Hattori 1989), reviewed by Sjoblom et al. (2008). These form a network which in transverse view resembles a basketweave or the so-called “small-square lattice” (Goldstein et al. 1990; Luther et al. 2002). The dense band of the Z-disc in longitudinal view has a precisely defined width that varies with fibre and muscle type: fast muscles have narrow widths (~30–50 nm), and slow and cardiac fibres have wide Z-bands (~100–140 nm; Luther et al. 2003; Rowe 1973). The widths are determined by the amount of overlap of the interdigitating actin filaments and by the number of Z-link layers which range from two in fast muscle (e.g. fish body white muscle) to six in slow muscle (Luther et al. 2003). The mechanism that maintains fixed Z-disc widths sometimes goes awry and the circumstances that can cause this as well as the consequences will be discussed.
Striated muscle sarcomere. a Schematic diagram showing the main components of the sarcomere. The A-band comprises myosin filaments crosslinked at the centre by the M-band assembly. Thin actin-containing filaments are tethered at their barbed end at the Z-disc and interdigitate with the thick filaments in the A-band. Two giant proteins contribute to the structure of the Z-disc. Nebulin (800 kDa) runs along the thin filaments and overlaps in the Z-disc (Pappas et al. 2008). The 3 MDa 1 μm long protein titin runs between the M-line and the Z-disc (Young et al. 1998). b Electron micrograph of a longitudinal section of fish white (fast) muscle showing details of the sarcomere. For the right-hand Z-disc, also reproduced partly in c, favourable alignment of the lattice gives the characteristic zigzag appearance, whereas for the left-hand Z-disc a more random lattice angle gives a blurred view. The measurement of the Z-disc width is quite ambiguous from these different appearances; hence one should measure it from the profile plot d which is derived from the Z-disc region in c. As defined in Luther (2000), the full width measured from near the base (lower horizontal arrows) is ~70 nm. A shoulder is typically seen in these plots (double head arrow) and this defines the region due to overlap of the actin filaments. The “overlap width” measured above the shoulder (upper horizontal arrows) is ~35 nm. These are important values to consider regarding the overlap of actin, titin and nebulin within the Z-disc (Pappas et al. 2008). Scale bar = 500 nm
Two giant polymer proteins, titin and nebulin/nebulette, like actin, also overlap within and form important parts of the Z-disc (Clark et al. 2002). Titin spans half sarcomeres between the M-band and Z-disc and forms the template for the sarcomere. It has important elastic regions (PEVK regions) in the I-band (Granzier and Labeit 2004; Tskhovrebova and Trinick 2003). Nebulin runs along the thin filament and forms the template for thin filament assembly (McElhinny et al. 2003). CapZ caps the barbed ends of the actin filaments and interacts strongly with α-actinin and nebulin (Papa et al. 1999; Pappas et al. 2008). The atomic structures of a few Z-disc components, including actin, α-actinin (from homologous domains), CapZ (Yamashita et al. 2003) and the titin-telethonin complex (Zou et al. 2006) have been solved recently and there is the exciting possibility of fitting them into high resolution 3D electron tomograms of the Z-disc in the near future. The filaments and proteins mentioned above will be discussed in detail in this review.
Passive transmission of tension through the Z-disc structural assembly is an important role of the Z-disc. However, the Z-disc has additional important roles as it houses or anchors an amazing number of additional proteins. These proteins have various roles, including involvement with stretch sensing and signalling (Epstein and Davis 2003; Pyle and Solaro 2004). Like all cells, muscle fibres and cardiomyocytes respond to stretch. For this response, stretch sensors are required which stimulate signalling proteins that eventually communicate with the nucleus. The identity and mechanism of the stretch sensors is not known, but they are believed to be located in the Z-disc and M-band (Knoll et al. 2002). Mutations in many of the Z-band proteins lead to disease. There have been excellent reviews recently on Z-disc proteins and their role in signalling and disease (Faulkner et al. 2001; Frank et al. 2006; Sheikh et al. 2007), so this topic will be discussed only briefly.
Reviews on the structure of the Z-disc include the comprehensive review written nearly 15 years ago by Vigoreaux (1994) and parts of sarcomere reviews since then (Clark et al. 2002; Craig and Padron 2004; Squire et al. 2005). There have been significant and exciting advances in our knowledge of the structure and function of the Z-disc in recent years and these form the main part of the current review.
Z-disc lead players
The structure and symmetry of the actin filament and of α-actinin are important factors for building a tetragonal lattice Z-disc in vertebrate striated muscles; interdigitating actin filaments and α-actinin form different number of layers of crosslinks between the antiparallel actin filaments in different Z-disc types. In insect flight muscle the rather different Z-disc is based on a hexagonal lattice (Deatherage et al. 1989) but it will not be discussed further here.
Structure of actin filaments
Actin is a ubiquitous, highly conserved, protein found in every eukaryotic cell. The 43 kD G-actin monomer is composed of 375 residues; they are identical between chicken and human and have only 39 differences with yeast actin (Sheterline et al. 1998). G-actin monomers polymerise to form filamentous F-actin and together with tropomyosin and the troponin complex, they form the muscle thin filaments. The atomic structure of G-actin was first solved by Kabsch et al. (1990) (Fig. 2a) and since then ~25 crystal structures have been solved (Reisler and Egelman 2007). The dimensions of the monomer are approximately 55 Å square (as viewed in Fig. 2a) by 35 Å deep. Four domains, numbered 1–4, have been identified on G-actin (Fig. 2a); domains 3 and 4 form the inner domain located closer to the filament axis. F-actin has not been crystallised, so the 3D structure has to be modelled from the G-actin atomic structure. The model by Holmes et al. (1990) based on X-ray fibre diffraction data and cryo-em 3D images has been widely accepted (Oda et al. 2009). Recently Oda et al. (2009) used higher resolution X-ray fibre diffraction data from highly aligned actin filament sols to model F-actin and found that the inner and outer domains of G-actin twist by 20° in the transition from G- to F-actin. The details of their modelling are shown in Fig. 2b–e. The Holmes model was built on 13/6 symmetry (13 G-actin monomers in 6 turns) which requires 166.2° rotation per monomer and is close to the symmetry of vertebrate muscle actin filaments. This results in a left-handed single stranded helix referred to as the genetic helix (follows the path 0, 1, 2 etc. in Fig. 2f). As the rotation per monomer is not far from 180°, the filament appears 2-stranded along a right handed long-pitched helix with a crossover repeat of 38 nm. The filament is highly inextensible axially (Egelman et al. 1982) with an axial rise per monomer of 27.5 Å. The rotation per monomer can vary (Egelman et al. 1982) and for insect flight muscle, in which the symmetry is 28/13, the rotation is 167.1° (Miller and Tregear 1972). This slightly different symmetry is also observed in the actin filament core in nemaline myopathy Z-bands (Morris et al. 1990) and slow muscle Z-bands (Luther and Squire 2002). Such symmetry is particularly favourable for Z-disc architecture since equivalent sites occur along the actin filament every seven monomers (~19 nm) with an exact rotation of 90°. This allows the construction of a Z-band in which interdigitating actin filaments arranged in square lattices are cross-linked with orthogonal layers of α-actinin every seven monomers (Fig. 2f).
Structure of G-actin and F-actin. a Crystal structure of G-actin (Kabsch et al. 1990). Four domains can be identified, labelled 1–4. Domains 3 and 4 lie closer to the centre of the actin filament helix. b–e Model of f-actin by Oda et al. (2009). They propose that G-actin undergoes a conformation change of 20° rotation of domain 2 upon incorporation into F-actin. b Model of F-actin using the transformed G-actin. The two monomers enclosed by an ellipse and the three monomers enclosed by a triangle are shown in detail in c and d, respectively, with the interacting residues listed. e Fitting of their model into the cryo-em 3D map of Narita et al. (2006). f Schematic diagram of F-actin. G-actin monomers pack along the single-stranded left-handed genetic helix (along monomers 0, 1, 2 etc.) or along two-stranded right-handed helices (along the red or green helix of monomers). In the Z-line and nemaline rods, actin filaments pack with 28/13 symmetry (28 actin monomers in 13 turns of genetic helix), with crossover length of 38.5 nm. Equivalent binding sites on the filament for Z-disc architecture occur every 19.2 nm, the span of seven monomers representing a half crossover, along actin with a rotation of 90°. These equivalent binding sites occur at monomers 0, 1, 7, 8, 14, 15 etc. (labelled with * or arrow for the hidden ones). a Reprinted by permission from Macmillian Publisher Ltd: Kabsch et al. (1990). b–e Reproduced by permission from Macmillan Publisher Ltd: Oda et al. (2009) (Color figure online)
Structure of α-actinin
Like actin, α-actinin is a ubiquitous protein found in eukaryotic cells [see reviews by Blanchard et al. (1989); Otey and Carpen (2004); Sjoblom et al. (2008)]. It belongs to the spectrin family of actin-binding proteins which include spectrin, dystrophin, utrophin and fimbrin. In non-muscle cells it is a principal component of stress fibres, adhesion plaques and attachment of the cytoskeleton to the membrane. In muscle, it forms the principal crosslinks at the Z-disc between actin filaments of opposite polarity originating from adjoining sarcomeres. It is a rod-shaped protein of length ~35 nm and occurs as an anti-parallel homodimer (Fig. 3a). The monomer comprises an NH2 terminal actin-binding domain (ABD) which is composed of two calponin homology domains. The C-terminus comprises two EF hand domains (calmodulin homology domains) that are Ca2+ sensitive in non-muscle isoforms. The central part of α-actinin comprises four tandem spectrin-like repeats each of which comprises a triple α-helix anti-parallel bundle (Fig. 3b). The ABD on each end of the dimer gives α-actinin the ability to crosslink actin filaments. So far each of the domains of α-actinin or one of the homologous proteins has been solved separately by X-ray crystallography (Djinovic-Carugo et al. 1999; Franzot et al. 2005; Ylanne et al. 2001), but the structure of the complete α-actinin monomer or dimer has not yet been determined at high resolution. Models of α-actinin have been made from the solved domains by fitting them to cryoelectron tomograms (Fig. 3b; Liu et al. 2004; Tang et al. 2001). The remarkable fact about the rod formed by the four spectrin repeats is that there is a twist of 90° from one end of the rod to the other (Ylanne et al. 2001). This, coupled with the flexible ABD, allows binding of α-actinin in a variety of conformations between antiparallel and parallel actin filaments and even along a single actin filament as shown recently by Hampton et al. (2007) in their study of α-actinin binding to actin filaments on lipid layers (Fig. 3c).
Structure of α-actinin. a α-actinin comprises N-terminal actin binding domains (ABD) that are calponin homology domains, four spectrin-like repeats (R1 to R4) comprising triple helical coiled-coils that form the rod domain and two EF hand domains that are calmodulin homology domains. b Shows a ribbon model of α-actinin depicted by Liu et al. (2004), PDB code 1sjj. c Demonstration of α-actinin binding modes on actin filaments by Hampton et al. (2007). Polarity of actin filament is marked with single or triple black arrowheads and α-actinin binds parallel and anti-parallel filaments. Binding of α-actinin along a single filament is marked with white arrowheads. Micrograph c from Hampton et al. (2007), with permission from Elsevier Science
The mode of α-actinin binding on actin was investigated by McGough et al. (1994) using cryo-electron microscopy. They showed that α-actinin binds over two neighbouring actin monomers along the long helix, namely with residues 348–355 in the first actin monomer and with residues 87–96 in the second. In their study of α-actinin binding to actin on lipid layers, Hampton et al. (2007) observed that the binding can occur at a range of angles, with a preference for 60 and 120°. The variation in binding angles may be due to varying torques, and the authors suggest that the variation in angle of attachment could be a tension sensing mechanism.
In addition to its structural role, it is now known that α-actinin has a major role in the docking of signalling and other proteins at the Z-disc (see reviews by Otey and Carpen 2004; Sjoblom et al. 2008).
Absence of tropomyosin and troponin from the Z-line
Tropomyosin and the troponin complex bind to the thin filament regularly over its entire length. Where does the binding start in relation to the Z-disc? This was investigated for troponin by Ohtsuki (1974). By applying troponin antibody to chicken breast (fast) muscle, he observed 24 stripes of spacing 38 nm over the full length of the thin filament (Fig. 4a). He reported the position of the first stripe at ~2 × 38 nm from the centre of the Z-band. The labelling pattern of tropomyosin was investigated by Trombitas et al. (1993). Using polyclonal antibodies against frog tibialis anterior (fast) muscle, they observed antibody labelling similar to Ohtsuki’s anti-troponin labelling (Fig. 4b). It appears from these studies that the binding of troponin and tropomyosin to thin filaments starts at 40–60 nm from the edge of the Z-band. Stromer and Goll (1972) reported that addition of tropomyosin and α-actinin to Z-line extracted fibres resulted in only α-actinin binding at the Z-line ends of the thin filaments.
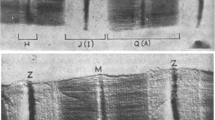
Electron micrographs showing that a troponin and b tropomyosin are not present in the Z-disk. a Troponin antibody labelled chicken breast muscle showing regular 38.5 nm stripes over the thin filaments; the first stripe occurs about 40 nm from the edge of the Z-disc (Ohtsuki 1974). b Tropomyosin antibody labelled frog semitendinosus muscle stretched to non-overlap shows the same periodicity stripes with the first stripe at about 60 nm from the edge of the Z-disc. Z-discs and an M-band are labelled Z and M. a From Ohtsuki (1974), with permission from Oxford. b From Trombitas et al. (1993), with permission from Springer. Scale bar = 0.5 μm
Structure of the Z-disc
Longitudinal section views
The Z-disc has characteristic appearances in longitudinal and transverse sections. The image in the electron microscope is a projected view of the structure through the depth of the sample. With different components at different depths in the section what is seen in the projected image can be quite blurred. In a typical section of ~100 nm thickness there are about 3–5 layers of unit cells of the Z-disc lattice (lattice size ~25 nm) within the depth of the section. Only if a section is oriented so that the appropriate lattice lines are aligned along the direction of view is the characteristic zigzag view of the Z-disc obtained (right side Z-disc in Fig. 1b and Fig. 7a). In most orientations, the appearance of the Z-disc resembles a fuzzy dense band (left side Z-band in Fig. 1b).
Transverse appearance
In transverse section electron micrographs there are two typical appearances of vertebrate muscle Z-bands, a distinct basketweave form and the so-called “small-square” form (Fig. 5). The origin of these views in relation to a unit cell of the Z-band comprising overlapping actin filaments is also shown (Fig. 5). Goldstein et al. (1987, 1988, 1990) and Yamaguchi et al. (1985) have proposed that the appearance of the Z-disc is a manifestation of its state. They suggested that the small-square form (Fig. 5a) occurs in relaxed muscle, but that during active contraction it transforms to the basketweave form (Fig. 5b) This is accompanied by a reduction in the lattice dimensions by 20%. Yamaguchi et al. (1985) showed how such a transition could occur by a computer simulation in which they varied the lattice dimensions. The idea is that in the basketweave form, a Z-link (α-actinin) spirals from one actin filament to the corresponding site 19 nm away axially on a neighbouring anti-parallel actin filament. In a transverse section micrograph, these spiral links provide the strands of the basketweave appearance. If instead of the spiral, the Z-links follows straighter paths and then change direction via acute bends, then a small-square lattice form is generated. 3D reconstructions of the small square Z-disc as found in nemaline myopathy (Morris et al. 1990) and of the basketweave form as found in fish fin muscle and bovine neck muscles (Luther 2000; Luther et al. 2002) support the proposed mechanism for the transformation.
Cross-sectional appearance of the Z-disc. a, b Demonstration by Goldstein and coworkers that the Z-disc in a muscle (here rat soleus muscle) cross-section can exist in a a small-square lattice form as in resting muscle or b a basketweave form when a muscle is activated (Goldstein et al. 1988). c, d Shematic drawings of the small-square and basketweave lattices with actin filament polarity depicted in green or red colours and showing the actin filament lattices extending on either side (Color figure online)
Early models of the Z-disc
The first 3D model of the Z-disc by Knappeis and Carlsen (1962) was derived from the observation that in longitudinal section electron micrographs actin filaments from two adjoining sarcomeres penetrate the Z-disc in staggered arrays and form zigzag links between their opposing terminal ends. Secondly, in transverse sections, interdigitating square lattices are seen with links between the opposing actin filaments. This observation led to the model in which four links from the end of one actin filament connect to the four neighbouring opposite polarity actin filaments from the adjoining sarcomere. In longitudinal view, this simple model would look the same along one lattice view or after rotation along the fibre axis by 90° (i.e. the [1, 0] and [0, 1] views, see Fig. 6). Luther (1991) showed that this is not the case and that the two views are quite different. The Knappeis and Carlsen model implies fourfold rotational symmetry which actin filaments do not possess. From their study of nemaline rod Z-bands, Morris et al. (1990) proposed that actin filaments have 43 screw symmetry. Another early Z-disc model was the looping filament model e.g. (Rowe 1973), but 3D reconstructions have found no evidence for them.
Nomenclature of Z-disc views. Actin filaments originating from two adjacent sarcomeres are coloured red and green. Electron micrographs represent projections of the structure in the depth of a section. The main lattice views are [1, 0] and [0, 1]. Halfway between these views is the [1, 1] view in which actin filaments in longitudinal sections appear continuous through the Z-disc. In this schematic model Z-links are not shown for clarity, hence the [1, 0] and [0, 1] views are the same. Adding Z-links to this model would make the two views very different (illustrated in the Z-disc models panel in Fig. 7). From Luther et al. (2003), with permission from Elsevier Science (Color figure online)
Variation of Z-disc structure with fibre-type
Vertebrate muscle Z-bands have precise widths that depend on fibre type (slow and fast) and muscle type (cardiac and skeletal). Fast muscles typically have narrow Z-bands and slow muscles have wide Z-bands. This is certainly an adaptation to fine tune the muscle to its function. Fast muscles have higher contraction velocities and produce high force. A detailed analysis of electron micrographs of the different types of Z-band was shown by Rowe (1973). The typical image of the Z-band in longitudinal sections is depicted as zigzag links between the opposing ends of actin filaments from adjacent sarcomeres, with a relative half unit cell shift between the two opposing arrays (Fig. 1). Rowe (1973) showed that the fast muscles had narrow Z-discs with a double chevron appearance and that slow muscles had wide Z-discs with 3 or 4 chevrons.
Z-disc widths have been shown to correlate with the number of layers of Z-link (α-actinin) connecting the anti-parallel actin filament ends that overlap in the Z-band. The numbers of layers identified so far are 2, 3, 4 and 6 (Luther et al. 2003). Note that as the image in the electron microscope is a projection of the density in the depth of a section, the zigzag connections in the Z-band do not directly give the number of Z-link/α-actinin layers. For example, a Z-disc showing a single zigzag layer (Fig. 1b) is actually due to two layers of α-actinin. To understand how the zigzag links arise, 3D reconstruction is required and this has been carried out for different Z-bands as described below. The results are summarised in Fig. 7 which shows for each Z-disc type a panel with the two orthogonal [1, 0] and [0, 1] views, a panel with the schematic views and a panel with the proposed 3D model showing the number of α-actinin links in that Z-disc.
Structure of vertebrate muscle Z-discs. The figure is organised into four rows, a, b, c & d showing respectively, 2-, 3-, 4- and 6-layer Z-bands. It is organised into three vertical panels, each panel with two images, comprising from the left, averaged electron micrographs, corresponding schematic views of structure and corresponding 3D models of the structure. The right half in each panel shows the appearance following 90° axial rotation. a Shows the Z-band in fish body muscle which comprises single zig-zag links between oppositely oriented actin filaments (Luther 1991). Modelling of 3D reconstructions suggest that that the Z-band is composed of two layers of α-actinin. This is the minimum width of a Z-band. By adding additional layers of α-actinin, we obtain b, c 3 and 4-layer Z-bands (also fast muscle), and d 6-layer Z-band from a slow muscle. The width is precisely maintained in each muscle type
3D structure of the Z-disc
Depicting the structure of the Z-disc on paper is difficult because the main components do not lie in a single plane. For example, in the [1, 0] lattice view actin filaments on either side of the Z-disc are arranged in different planes. Hence the schematic longitudinal view of a zigzag link between actin filaments from adjoining sarcomeres is a composite (projection) as the layers of actin filaments from adjacent sarcomeres and the Z-links occur in different planes. The Z-links are composed mainly of α-actinin, but other proteins may contribute like nebulin, titin and myotilin. The nomenclature for describing the different projections is shown in Fig. 6. The main lattice views are referred to as the [1, 0] and [0, 1] views or projections.
3D structure of the 2-layer Z-disc: the building block of vertebrate Z-discs
Franzini-Armstrong (1973) first reported a very narrow Z-band in fish white muscle which is a fast muscle. She noted that in certain orientations in longitudinal sections, the actin filaments ends of two adjacent sarcomeres were connected by a single zigzag link (Fig. 1b). Luther (1991) undertook a detailed 3D study of this muscle and showed that the zigzag appearance was one of the two main lattice views and that rotating the Z-band by 90° along the myofibril axis gave a different view that resembled overlapping spikes (Fig. 7a). We note that the image is symmetrical about the centre of the Z-band (strictly there is glide plane symmetry relating the upper and lower halves. The actin filaments from the adjacent sarcomeres were found to be cross-linked with two pairs of orthogonal links separated along the actin filament by ~10 nm (Luther 1991). This is probably 19 nm in light of new data described below.
Luther also described “polar” links at the periphery of the Z-band between actin filaments of the same sarcomere. Such links have been reported in other studies (Schroeter et al. 1996; Trombitas et al. 1988; Tskhovrebova 1991). As described in the section on α-actinin, Taylor and colleagues have shown that α-actinin can crosslink polar as well as bipolar actin filaments (see Fig. 3).
3D structure of a 3-layer Z-disc
A 3-layer Z-disc was first observed in fish fin muscle (Luther 2000). The two orthogonal views are shown in Fig. 7b. The appearance comprises one zigzag layer and bulbous densities which occur on different sides in the orthogonal [1, 0] and [0, 1] views. Unlike the symmetrical appearance of the simple 2-layer Z-disc relating the upper and lower halves of the Z-disc (Fig. 7a), the 3-layer has quite different appearances. This difference occurs because of the underlying even and odd number of layers of Z-links. Hence, for an unknown Z-band, one can determine whether it comprises even or odd layers by looking for a symmetrical appearance relating the Z-disc parts in adjoining sarcomeres.
3D structure of a 4-layer Z-disc
A formal 3D reconstruction of a 4 layer Z-disc has not been done. Examination of frog sartorius (fast) muscle Z-disc in longitudinal sections (Luther et al. 2003) revealed Z-discs between two sarcomeres with regions of two different widths. The narrow one had the same pattern as the three layer Z-disc. The second one, wider by ~18 nm, had symmetrical appearance about the centre of the Z-disc and was therefore inferred to comprise four layers (Fig. 7c).
3D structure of a 6-layer Z-disc
Slow and cardiac muscles typically have wide Z-discs, ~100–140 nm wide (Fig. 7d). The striking difference between the orthogonal lattice views is that in one of them, e.g. the [1, 0] view, the centre has distinct bars, whereas the [0, 1] view has a fuzzy region. The 3D reconstruction of a wide Z-disc from bovine neck slow muscle (Luther et al. 2002) was found to comprise a 6-layer Z-disc. With 6 layers of α-actinin crosslinking the overlapping actin filaments from adjacent sarcomeres this must form a relatively rigid Z-disc that should be most resistant to distortion during muscular activity.
3D structure of the nemaline rod Z-band
Nemaline myopathy is a congenital disease of skeletal muscles characterized by muscle weakness (Wallgren-Pettersson and Laing 2006). It is characterised histologically by granules revealed by gomori trichrome stain. In the electron microscope it presents as enlarged Z-bands within the striated fibres and isolated ~1 μm long nemaline rods (Fig. 8a). Nemaline myopathy arises from mutations in several proteins which include actin and those related to actin, including nebulin, tropomyosin and troponin-T (Wallgren-Pettersson and Laing 2006). Interestingly, nemaline myopathy does not appear to be associated with mutations in α-actinin or titin.
Structure of the nemaline rod Z-band. a Nemaline myopathy is characterised by regions of enlarged irregular Z-discs and large isolated nemaline rods. b–e 3D reconstruction of the nemaline Z-band (Morris et al. 1990). A cross-sectional view of the reconstruction is depicted as stereo chicken-wire b and a solid model c which shows actin filaments of opposite polarity (labelled A, B), and crosslinks through an axial strut C. d, e Show the same views in a longitudinal section. A model for the mode of crosslinking is shown in f with actin filament polarity depicted by arrows above and below. The path of two α-actinin molecules shares a central axial region. This model represents the small-square lattice structure. b–f From Morris et al. (1990), with permission from Rockefeller University Press. Scale bar (a) = 0.5 μm
The 3D structure of the nemaline rod Z-band was investigated by Morris et al. (1990) in a classic paper in Z-band research. Stereo views of the 3D reconstruction and a schematic view of the model are shown in Fig. 8. As the appearance of nemaline rods in transverse view is the small-square type, this reconstruction gives insight into the architecture of such Z-discs. Morris et al. found that the crosslink comprises three parts, a central part that runs axially and two perpendicular parts that link neighbouring antiparallel filaments (Fig. 8f). They proposed that the axial part is composed of the rod part of two α-actinin molecules which link across to four actin filaments. Recently myotilin has been identified as a component of nemaline rods (Schroder et al. 2003), so there may be more components to fit in the 3D architecture of the nemaline and other Z-bands.
Loss of fixed-width in the Z-disc
As mentioned before, vertebrate striated muscle Z-discs have precisely defined widths for particular muscle types. To date we have identified widths that correspond to 2, 3, 4 and 6 layers of Z-link/α-actinin. There may be other Z-discs of different number of layers yet to be found. Z-bands with patches of two different widths within a single sarcomere were reported by Luther et al. (2003) and they were identified as 3 and 4 layer Z-bands. The cause for the variation within a single sarcomere is not certain but it may be a snapshot of transition of a fibre from one type to another.
Loss of Z-band width definition occurs in a variety of circumstances. As discussed earlier, nemaline myopathy is characterised by large isolated nemaline rods, but it also has regions of striated fibres with irregular width Z-bands (Fig. 8). In mice muscles deficient of muscle LIM protein (MLP; description later), the Z-bands in cardiac muscle were frequently irregular and widened (Knoll et al. 2002). Electron microscopy of human cardiac tissue from myectomy operations of hypertrophic cardiomyopathy patients sometimes shows irregular Z-bands that are often spindle shaped (author’s unpublished observations). Since cardiac patients are frequently quite old, we may ask whether the Z-disc defects are due to cardiac pathology or due to old age. Examination of the hearts in very old mice (nearly 2 years) has shown scattered regions with irregular or spindle shaped Z-discs (author’s unpublished observations). Zolk et al. (2000) have reported reduced levels of MLP in failing hearts and this reduction of MLP may also be the cause of irregular Z-bands observed in very old mice.
Massive loss of Z-disc integrity occurs following muscle injury (Jones et al. 2004; Lieber and Friden 2002). It is characterised by Z-line “streaming” in which Z-disc structure is disrupted and Z-disc material appears ripped out across a large part of a sarcomere. This occurs particularly after eccentric exercise. Z-line streaming is also common in several muscle disorders (Goebel 2002).
Important structural proteins in the Z-disc
Nebulin/nebulette
Nebulin is an 800 kDa protein that runs along the length of the thin filaments (Labeit and Kolmerer 1995; Wang and Wright 1988). Although not fully understood, nebulin plays an important role in the assembly, structure and function of the Z-disc in skeletal muscle (McElhinny et al. 2003). In particular it may be that nebulin helps to define the thin filament length in many skeletal muscles (Littlefield and Fowler 2008; Pappas et al. 2008; Witt et al. 2006). In cardiac muscle there may be a miniscule amount of nebulin or possibly none (McElhinny et al. 2003) and this may be related to the variable lengths of thin filaments in cardiac muscle (Burgoyne et al. 2008). Nebulette is a 107 kd nebulin homologue present in the cardiac muscle Z-disc (Moncman and Wang 1995). During myofibrillogenesis nebulin appears in I-Z-I bodies, precursors of Z-discs, before thin filaments attain full length (McElhinny et al. 2003). Nebulin is composed of repeating 35-residue α-helical domains with the conserved motif SDxxYK (Labeit et al. 1991) labelled M1 to M185. Nebulin runs along the full length of the thin filaments with the C-terminus located in the Z-disc. A single nebulin module interacts with each actin monomer along the actin filament (Pfuhl et al. 1994). Along most of its length, the repeating modules are organised into super-repeats of seven domains and this is thought to reflect the binding of tropomyosin and troponin to the thin filament (Labeit and Kolmerer 1995). The super-repeat is absent from the 100 kDa region from module M162 to the C-terminus. Previously it was thought that nebulin terminated at the near edge of the Z-band and did not overlap the Z-disc width (Millevoi et al. 1998). However, a new study by Pappas et al. (2008) on chick skeletal myotubes showed that nebulin is required for the localisation of CapZ in the Z-disc; they found that domains M160 to M164 bind to CapZ and that the remainder of the C-terminus interacts with the Z-band. Therefore, they proposed a model in which a nebulin molecule binds along thin filaments up to the near Z-disc boundary, then moves to the end of a neighbouring antiparallel actin filament where it interacts with its CapZ terminus. It then continues along this filament to the distal edge of the Z-disc (Fig. 1a). As a consequence nebulin has the potential to contribute to the tension bearing role of the Z-disc.
The effect of nebulin depletion has been investigated by two groups (Bang et al. 2006; Witt et al. 2006). Nebulin-knockout mice were growth retarded at birth and 90% died within 2 weeks. The mice had normal levels of CapZ, but the myopalladin levels (another Z-disc component) were much reduced. Both groups found that the thin filaments were shorter by 15–20%. Witt et al. found no nebulin in cardiac muscle, contradicting suggestions that there may be a small amount present that regulates the thin filament length in some fashion. Both groups observed widened Z-bands and the nemaline rod phenotype. They suggested that impaired thin filament capping contributes to a severe nemaline myopathy pathology resulting from thin filaments growing beyond the normal Z-width.
Titin
Titin is a giant 3 MDa protein that spans half sarcomeres from the Z-disc to the M-band [see reviews (Granzier and Labeit 2004; Linke 2008)]. The C- and N-terminii span the full width of the M-band and Z-disc, hence consecutive titins form a continuous structure from one end of the sarcomere to the other. The N-terminus 90 kDa of titin is located at the Z-disc (Gregorio et al. 1998; Young et al. 1998). Of this, the N-terminal 30 kDa region comprising the Ig domains Z1 and Z2 is located at the distal end of the Z-disc and the 60 kDa region comprising the so-called Z-repeats spans the width of the Z-disc (Gregorio et al. 1998).
The Z-repeats are 45 residue modules (Gautel et al. 1996). Labelled Zr1 to Zr7, they are differentially expressed in different muscle or fibre types, with fast muscle having only two Z-repeats (Ohtsuka et al. 1997) or four Z-repeats (Sorimachi et al. 1997), slow muscle having six repeats and cardiac muscle seven repeats (Gautel et al. 1996; Sorimachi et al. 1997). Gautel et al. (1996) and Young et al. (1998) proposed that the Z-repeats may form the template for Z-disc assembly with each Z-repeat contributing to a layer of α-actinin. This requires that the width of each Z-repeat match the periodicity of α-actinin layers in the Z-band. However, Luther and Squire (2002) showed that the periodicity within the Z-disc is nearly 19 nm. They argued that since the length of a Z-repeat is much less than 19 nm (Atkinson et al. 2000), a single Z-repeat cannot act as a template, but two Z-repeats could have the right dimensions (Fig. 9). It should be noted here that perhaps there is no need for an external template for the assembly of the Z-disc. The template for Z-disc assembly is surely provided by the symmetry of actin filaments, the arrangement of parallel and anti-parallel filaments in a tetragonal lattice and the flexible binding sites at each end of the homodimeric α-actinin. Indeed, this is how nemaline rods appear to form spontaneously following mutations or physiological insult e.g. following tenotomy (Yamaguchi et al. 1983). What is special about the striated muscle Z-disc is its fixed width in particular types of muscles, so it is essential to have markers that define the limits of Z-disc assembly. The start and end markers may be provided by a combination of Z-repeats and the Z-disc region of nebulin which is also differentially expressed in different fibre types (Pappas et al. 2008; Witt et al. 2006; Young et al. 1998).
Mismatch of titin Z-repeats and Z-disc periodicity. The figure shows a scale drawing of two oppositely oriented actin filaments (A and B) in a 6-layer Z-band (a) against the Z-band region of titin comprising Z-repeats, Zr (b). The dimensions of Z-repeats need to match the Z-band periodicity of ~19 nm in order to act as template for different Z-bands as proposed by Young et al. (1998). The Z-repeats are probably much shorter and two Z-repeats as shown in (c) may form the template. From Luther and Squire (2002), with permission from Elsevier Science
Zou et al. (2006) have crystallised and solved a complex of a pair of titin Z1/Z2 domains sandwiching a telethonin (T-cap) molecule. They showed a fascinating structure comprising a palindromic arrangement in which the Z1/Z2 domains originating from the same actin filament are glued by telethonin (Fig. 10a–c). In subsequent dynamic simulations, the authors show that the interaction holding the domains together is strong (Lee et al. 2006). They propose that this is necessary for the strong forces experienced by the titin molecule. However, it is currently not clear what interacting partners are present that could pull apart the two titin molecules in the palindromic partnership. Clearly there is a need for more detailed 3D models of the Z-disc that may identify any binding partner to the Z1/Z2 telethonin complex.
Crystal structures of components of the Z-disc. a, b Front and back views of two titin Z1–Z2 modules (light and dark blue) sandwiched with a telethonin molecule (red), forming a strongly bound palindromic complex. Zou et al. (2006) propose a model c in which two titin molecules associated with an actin filament form the palindromic complex and interact with MLP, FATZ etc. d Cryo-em determination of CapZ binding to the barbed end of actin filaments, as expected at the Z-band. Narita et al. (2006) propose the mechanism for the CapZ binding the terminal two actin monomers via the β- and α-tentacles. a–c From Zou et al. (2006), with permission from Nature Publishing Group. d From Narita et al. (2006), with permission from Nature Publishing Group (Color figure online)
CapZ
CapZ is a heterodimer that binds tightly and caps the barbed ends of actin filaments in the Z-disc. It also strongly binds a spectrin domain of α-actinin (Papa et al. 1999) and the C-terminus (domain M162) of nebulin (Pappas et al. 2008). The crystal structure of CapZ (Yamashita et al. 2003) showed the arrangement of the α and β subunits. The C-terminal domains (~30 residues) of both subunits have α-helical regions and Yamashita et al. (2003) propose that these regions act like tentacles for binding actin. To determine the 3D localisation of CapZ on actin filaments, Narita et al. (2006) undertook cryo-electron microscopy of CapZ-bound actin filaments. Their 3D images at 2 nm resolution show distinct features of CapZ. From the known atomic structures of CapZ and the actin filament, they modelled the binding on their cryo-em maps (Fig. 10d).
Z-band proteins involved in signalling and disease
For just over a decade, the Z-band has been a treasure trove for the biochemist/geneticist, with a multitude of new proteins being discovered. These have been exciting discoveries as mutations in the genes for these proteins lead to muscle diseases and cardiomyopathies. Some of the proteins have domains that are well known in biology, like the PDZ and LIM domains, so their importance was immediately realised. Excellent reviews on these proteins and their role in signalling and disease have been written (Clark et al. 2002; Faulkner et al. 2001; Frank et al. 2006; Pyle and Solaro 2004; Sheikh et al. 2007) and only a brief summary is included here.
For the newcomer it is a daunting prospect to understand all the new proteins. One could classify them by their binding partners; for example, several proteins bind α-actinin. However, the proteins have extensive interactions with each other and as Faulkner et al. (2001) state, they are not “islands unto themselves”. These interactions are illustrated schematically in the reviews above and in the review by Lange et al. (2006).
In a recent study, some of the Z-band proteins have been classified into three families, myotilin, FATZ and enigma (von Nandelstadh et al. 2009, and references therein). The myotilin family includes myotilin, palladin and myopalladin. These proteins have immunoglobulin domains and bind α-actinin, filamin and FATZ. The FATZ family includes FATZ (also called casarcin and myozenin) and their binding partners include myotilin, filamin, telethonin, α-actinin and ZASP. FATZ-1 and FATZ-3 occur in fast twitch muscles and FATZ-2 occurs in slow twitch and cardiac fibres. The enigma family of proteins is characterized by an NH2-terminal PDZ domain and nought to three LIM domains at the COOH terminal. PDZ domains are structural domains of 80–90 amino acids arranged in 5–6 β-strands and 2 α-helices and are involved in protein–protein interactions (Harris and Lim 2001). LIM domains are protein interaction domains of about 55 amino acids (Kadrmas and Beckerle 2004). They are composed of tandem zinc finger domains connected by a hydrophobic linker of two amino acids. Cypher/ZASP/Oracle is probably the most studied enigma member (Faulkner et al. 1999; Passier et al. 2000; Zhou et al. 2001). Cypher/ZASP may serve as a linker-strut by binding to α-actinin via its PDZ domain and may be involved in signalling as it binds protein kinase C via its LIM domains. Cypher-deficient mice die soon after birth and analysis of contracting and non-contracting (diaphragm) muscles suggests that cypher is essential for maintaining Z-band structure and muscle integrity (Zhou et al. 2001). Mutations in cypher lead to dilated cardiomyopathy and muscle myopathies now termed Zaspopathies.
Myofibrillar myopathy (MFM) is a muscle disorder especially relevant to the Z-disc. MFM is a morphologically distinct group of muscle pathologies of skeletal and cardiac muscle characterised by disintegration of the Z-disc and abnormal accumulation of proteins (Selcen and Engel 2004). It is caused by mutations in the genes for proteins involved in maintaining the structural integrity of the Z-disc. To date, these include desmin, αB-crystallin, myotilin and ZASP.
Muscle LIM protein (MLP) is a highly researched LIM only protein that has several binding partners including telethonin, α-actinin, Myo-D, N-RAP and β-spectrin (Gehmlich et al. 2008; Knoll et al. 2002). The MLP-telethonin-titin interaction was thought to be involved in mechanotransduction i.e. to be the elusive stretch sensor mechanism (Knoll et al. 2002). New research by Geier et al. (2008) using monoclonal antibodies against MLP suggest that MLP may not have specific a Z-band location as previously thought but has diffuse cytoplasmic location. Hence Gehmlich et al. (2008) suggest that MLP may be a downstream signal transducer in mechano-signalling cascades. The search for the stretch-sensor mechanism continues.
Future directions
The image of the Z-disc as a sarcomere boundary marker passively involved in contraction has now moved more centre stage to that of a control watch tower. Teeming with proteins that can interact with each other, proteins with domains with established roles in biology for signalling and protein–protein interactions, the Z-disc is a fascinating assembly. There are important questions to address. What is the basis of the stretch sensing mechanism? What is the mechanism that goes awry in aging and disease that leads to loss of a fixed width in the Z-disc? Urgently needed now are high resolution tomographic studies that can identify the major members, actin and α-actinin, identify the recently solved structures of the titin Z1/Z2-telethonin complex and of CapZ and finally attempt to identify new members like myotilin and myopalladin.
References
Atkinson RA, Joseph C, Dal Piaz F, Birolo L, Stier G, Pucci P, Pastore A (2000) Binding of alpha-actinin to titin: implications for Z-disk assembly. Biochemistry 39:5255–5264
Bang ML, Li X, Littlefield R, Bremner S, Thor A, Knowlton KU, Lieber RL, Chen J (2006) Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J Cell Biol 173:905–916
Blanchard A, Ohanian V, Critchley D (1989) The structure and function of alpha-actinin. J Muscle Res Cell Motil 10:280–289
Burgoyne T, Muhamad F, Luther PK (2008) Visualization of cardiac muscle thin filaments and measurement of their lengths by electron tomography. Cardiovasc Res 77:707–712
Clark KA, McElhinny AS, Beckerle MC, Gregorio CC (2002) Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol 18:637–706
Craig R, Padron R (2004) Molecular structure of the sarcomere. In: Engel AC, Franzini-Armstrong C (eds) Myology, 3rd edn. McGraw-Hill, New York, p 129
Deatherage JF, Cheng NQ, Bullard B (1989) Arrangement of filaments and cross-links in the bee flight muscle Z disk by image analysis of oblique sections. J Cell Biol 108:1775–1782
Djinovic-Carugo K, Young P, Gautel M, Saraste M (1999) Structure of the alpha-actinin rod: molecular basis for cross-linking of actin filaments. Cell 98:537–546
Egelman EH, Francis N, DeRosier DJ (1982) F-actin is a helix with a random variable twist. Nature 298:131–135
Epstein ND, Davis JS (2003) Sensing stretch is fundamental. Cell 112:147–150
Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, Valle G, Lanfranchi G (1999) ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol 146:465–475
Faulkner G, Lanfranchi G, Valle G (2001) Telethonin and other new proteins of the Z-disc of skeletal muscle. IUBMB Life 51:275–282
Frank D, Kuhn C, Katus HA, Frey N (2006) The sarcomeric Z-disc: a nodal point in signalling and disease. J Mol Med 84:446–468
Franzini-Armstrong C (1973) The structure of a simple Z line. J Cell Biol 58:630–642
Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K (2005) The crystal structure of the actin binding domain from alpha-actinin in its closed conformation: structural insight into phospholipid regulation of alpha-actinin. J Mol Biol 348:151–165
Gautel M, Goulding D, Bullard B, Weber K, Furst DO (1996) The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J Cell Sci 109(Pt 11):2747–2754
Gehmlich K, Geier C, Milting H, Furst D, Ehler E (2008) Back to square one: what do we know about the functions of Muscle LIM Protein in the heart? J Muscle Res Cell Motil 29:155–158
Geier C, Gehmlich K, Ehler E, Hassfeld S, Perrot A, Hayess K, Cardim N, Wenzel K, Erdmann B, Krackhardt F, Posch MG, Osterziel KJ, Bublak A, Nagele H, Scheffold T, Dietz R, Chien KR, Spuler S, Furst DO, Nurnberg P, Ozcelik C (2008) Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet 17:2753–2765
Goebel HH (2002) Central core disease. In: Karpati G (ed) Structural and molecular basis of skeletal muscle diseases. ISN Neuropath Press, Basel, pp 65–67
Goldstein MA, Michael LH, Schroeter JP, Sass RL (1987) Z band dynamics as a function of sarcomere length and the contractile state of muscle. FASEB J 1:133–142
Goldstein MA, Michael LH, Schroeter JP, Sass RL (1988) Structural states in the Z band of skeletal muscle correlate with states of active and passive tension. J Gen Physiol 92:113–119
Goldstein MA, Schroeter JP, Sass RL (1990) Two structural states of the vertebrate Z band. Electron Microsc Rev 3:227–248
Granzier HL, Labeit S (2004) The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res 94:284–295
Gregorio CC, Trombitas K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, Sorimachi H, Labeit S (1998) The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J Cell Biol 143:1013–1027
Hampton CM, Taylor DW, Taylor KA (2007) Novel structures for alpha-actinin: F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J Mol Biol 368:92–104
Harris BZ, Lim WA (2001) Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci 114:3219–3231
Holmes KC, Popp D, Gebhard W, Kabsch W (1990) Atomic model of the actin filament. Nature 347:44–49
Jones D, Round J, de Haan A (2004) Skeletal muscle from molecules to movement. Churchill Livingstone, Edinburgh. ISBN 0443074275
Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC (1990) Atomic structure of the actin: DNase I complex. Nature 347:37–44
Kadrmas JL, Beckerle MC (2004) The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 5:920–931
Knappeis GG, Carlsen F (1962) The ultrastructure of the Z disc in skeletal muscle. J Cell Biol 13:323–335
Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR (2002) The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111:943–955
Labeit S, Kolmerer B (1995) The complete primary structure of human nebulin and its correlation to muscle structure. J Mol Biol 248:308–315
Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, Wardale J, Trinick J (1991) Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett 282:313–316
Lange S, Ehler E, Gautel M (2006) From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol 16:11–18
Lee EH, Gao M, Pinotsis N, Wilmanns M, Schulten K (2006) Mechanical strength of the titin Z1Z2-telethonin complex. Structure 14:497–509
Lieber RL, Friden J (2002) Mechanisms of muscle injury gleaned from animal models. Am J Phys Med Rehabil 81:S70–S79
Linke WA (2008) Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res 77:637–648
Littlefield RS, Fowler VM (2008) Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Semin Cell Dev Biol 19:511–519
Liu J, Taylor DW, Taylor KA (2004) A 3-D reconstruction of smooth muscle alpha-actinin by CryoEm reveals two different conformations at the actin-binding region. J Mol Biol 338:115–125
Luther PK (1991) Three-dimensional reconstruction of a simple Z-band in fish muscle. J Cell Biol 113:1043–1055
Luther PK (2000) Three-dimensional structure of a vertebrate muscle Z-band: implications for titin and alpha-actinin binding. J Struct Biol 129:1–16
Luther PK, Squire JM (2002) Muscle Z-band ultrastructure: titin Z-repeats and Z-band periodicities do not match. J Mol Biol 319:1157–1164
Luther PK, Barry JS, Squire JM (2002) The three-dimensional structure of a vertebrate wide (slow muscle) Z-band: lessons on Z-band assembly. J Mol Biol 315:9–20
Luther PK, Padron R, Ritter S, Craig R, Squire JM (2003) Heterogeneity of Z-band structure within a single muscle sarcomere: implications for sarcomere assembly. J Mol Biol 332:161–169
McElhinny AS, Kazmierski ST, Labeit S, Gregorio CC (2003) Nebulin: the nebulous, multifunctional giant of striated muscle. Trends Cardiovasc Med 13:195–201
McGough A, Way M, DeRosier D (1994) Determination of the alpha-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol 126:433–443
Miller A, Tregear RT (1972) Structure of insect fibrillar flight muscle in the presence and absence of ATP. J Mol Biol 70:85–104
Millevoi S, Trombitas K, Kolmerer B, Kostin S, Schaper J, Pelin K, Granzier H, Labeit S (1998) Characterization of nebulette and nebulin and emerging concepts of their roles for vertebrate Z-discs. J Mol Biol 282:111–123
Moncman CL, Wang K (1995) Nebulette: a 107 kD nebulin-like protein in cardiac muscle. Cell Motil Cytoskeleton 32:205–225
Morris EP, Nneji G, Squire JM (1990) The three-dimensional structure of the nemaline rod Z-band. J Cell Biol 111:2961–2978
Narita A, Takeda S, Yamashita A, Maeda Y (2006) Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J 25:5626–5633
Oda T, Iwasa M, Aihara T, Maeda Y, Narita A (2009) The nature of the globular- to fibrous-actin transition. Nature 457:441–445
Ohtsuka H, Yajima H, Maruyama K, Kimura S (1997) The N-terminal Z repeat 5 of connectin/titin binds to the C-terminal region of alpha-actinin. Biochem Biophys Res Commun 235:1–3
Ohtsuki I (1974) Localization of troponin in thin filament and tropomyosin paracrystal. J Biochem 75:753–765
Otey CA, Carpen O (2004) Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 58:104–111
Papa I, Astier C, Kwiatek O, Raynaud F, Bonnal C, Lebart MC, Roustan C, Benyamin Y (1999) Alpha actinin-CapZ, an anchoring complex for thin filaments in Z-line. J Muscle Res Cell Motil 20:187–197
Pappas CT, Bhattacharya N, Cooper JA, Gregorio CC (2008) Nebulin interacts with CapZ and regulates thin filament architecture within the Z-disc. Mol Biol Cell 19:1837–1847
Passier R, Richardson JA, Olson EN (2000) Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev 92:277–284
Pfuhl M, Winder SJ, Pastore A (1994) Nebulin, a helical actin binding protein. EMBO J 13:1782–1789
Pyle WG, Solaro RJ (2004) At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res 94:296–305
Reisler E, Egelman EH (2007) Actin structure and function: what we still do not understand. J Biol Chem 282:36133–36137
Rowe RW (1973) The ultrastructure of Z disks from white, intermediate, and red fibers of mammalian striated muscles. J Cell Biol 57:261–277
Schroder R, Reimann J, Salmikangas P, Clemen CS, Hayashi YK, Nonaka I, Arahata K, Carpen O (2003) Beyond LGMD1A: myotilin is a component of central core lesions and nemaline rods. Neuromuscul Disord 13:451–455
Schroeter JP, Bretaudiere JP, Sass RL, Goldstein MA (1996) Three-dimensional structure of the Z band in a normal mammalian skeletal muscle. J Cell Biol 133:571–583
Selcen D, Engel AG (2004) Mutations in myotilin cause myofibrillar myopathy. Neurology 62:1363–1371
Sheikh F, Bang ML, Lange S, Chen J (2007) “Z”eroing in on the role of cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc Med 17:258–262
Sheterline P, Clayton J, Sparrrow JC (1998) Actin. Oxford University Press, Oxford
Sjoblom B, Salmazo A, Djinovic-Carugo K (2008) Alpha-actinin structure and regulation. Cell Mol Life Sci 65:2688–2701
Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S (1997) Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol 270:688–695
Squire JM, Al-Khayat HA, Knupp C, Luther PK (2005) Molecular architecture in muscle contractile assemblies. Adv Protein Chem 71:17–87
Stromer MH, Goll DE (1972) Studies on purified alpha-actinin. II. Electron microscopic studies on the competitive binding of alpha-actinin and tropomyosin to Z-line extracted myofibrils. J Mol Biol 67:489–494
Takahashi K, Hattori A (1989) Alpha-actinin is a component of the Z-filament, a structural backbone of skeletal muscle Z-disks. J Biochem (Tokyo) 105:529–536
Tang J, Taylor DW, Taylor KA (2001) The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca(2 +)-dependent actin binding. J Mol Biol 310:845–858
Trombitas K, Baatsen PH, Pollack GH (1988) I-bands of striated muscle contain lateral struts. J Ultrastruct Mol Struct Res 100:13–30
Trombitas K, Frey L, Pollack GH (1993) Filament lengths in frog semitendinosus and tibialis anterior muscle fibres. J Muscle Res Cell Motil 14:167–172
Tskhovrebova LA (1991) Vertebrate muscle Z-line structure: an electron microscopic study of negatively-stained myofibrils. J Muscle Res Cell Motil 12:425–438
Tskhovrebova L, Trinick J (2003) Titin: properties and family relationships. Nat Rev Mol Cell Biol 4:679–689
Vigoreaux JO (1994) The muscle Z band: lessons in stress management. J Muscle Res Cell Motil 15:237–255
von Nandelstadh P, Ismail M, Gardin C, Suila H, Zara I, Belgrano A, Valle G, Carpen O, Faulkner G (2009) A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol Cell Biol 29:822–834
Wallgren-Pettersson C, Laing NG (2006) 138th ENMC workshop: nemaline myopathy, 20–22 May 2005, Naarden, The Netherlands. Neuromuscul Disord 16:54–60
Wang K, Wright J (1988) Architecture of the sarcomere matrix of skeletal muscle: immunoelectron microscopic evidence that suggests a set of parallel inextensible nebulin filaments anchored at the Z line. J Cell Biol 107:2199–2212
Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S (2006) Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J 25:3843–3855
Yamaguchi M, Robson RM, Stromer MH, Cholvin NR, Izumimoto M (1983) Properties of soleus muscle Z-lines and induced Z-line analogs revealed by dissection with Ca2 + -activated neutral protease. Anat Rec 206:345–362
Yamaguchi M, Izumimoto M, Robson RM, Stromer MH (1985) Fine structure of wide and narrow vertebrate muscle Z-lines. A proposed model and computer simulation of Z-line architecture. J Mol Biol 184:621–643
Yamashita A, Maeda K, Maeda Y (2003) Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J 22:1529–1538
Ylanne J, Scheffzek K, Young P, Saraste M (2001) Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure (Camb) 9:597–604
Young P, Ferguson C, Banuelos S, Gautel M (1998) Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of alpha-actinin. EMBO J 17:1614–1624
Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, Knoll G, Shelton GD, Evans S, Chen J (2001) Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol 155:605–612
Zolk O, Caroni P, Bohm M (2000) Decreased expression of the cardiac LIM domain protein MLP in chronic human heart failure. Circulation 101:2674–2677
Zou P, Pinotsis N, Lange S, Song YH, Popov A, Mavridis I, Mayans OM, Gautel M, Wilmanns M (2006) Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature 439:229–233
Acknowledgments
Much of the work I have done on the sarcomere has been in collaboration with John Squire and I am greatly indebted to him. I have also collaborated with and had extensive discussions on the structure of the Z-disc with Ed Morris, John Barry, Tom Burgoyne, Roger Craig and Raul Padron. I would like to thank Cathy Timson for excellent technical help. I am grateful to John Squire, Roger Craig, Georgine Faulkner and Larissa Tskhovrebova for invaluable comments on the manuscript. I am grateful to the British Heart Foundation (Project PG/06/010) for their support of my research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s10974-010-9236-3.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Luther, P.K. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J Muscle Res Cell Motil 30, 171–185 (2009). https://doi.org/10.1007/s10974-009-9189-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-009-9189-6