Abstract
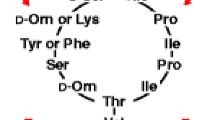
Antimicrobial peptides (AMPs) are natural antagonistic tools of many bacteria and are considered as attractive antimicrobial agents for the treatment of bacteria with multidrug resistance. Lactic acid bacteria from the gastrointestinal tract of animals and human produce various AMPs inhibiting the growth of pathogens. Here we report the isolation and identification of novel Lactobacillus fermentum strain HF-D1 from the human gut producing AMPs which prevents the growth of P. aeruginosa and S. marcescens. The active fraction of peptides was obtained from the culture liquid by precipitation at 80% saturation of ammonium sulphate. For peptides identification, the precipitate was treated with guanidine hydrochloride to desorb from proteins, separated with ultrafiltration on spin columns with 10,000 MWCO, desalted with a reversed-phase chromatography and subjected to LC–MS/MS analysis. The in silico analysis of the identified 1111 peptides by using ADAM, CAMPR3 and AMPA prediction servers led to identification of the linear peptide with highly probable antimicrobial activity and further investigation of its antibacterial activity mechanism is promising. By using the dereplication algorithm, the peptide highly similar to non-ribosomal cyclic AMPs originally isolated from Staphylococcus epidermidis has been identified. This indicates that L. fermentum HF-D1 represents a novel strain producing antimicrobial peptides targeting P. aeruginosa and S. marcescens.



Similar content being viewed by others
Abbreviations
- ADAM:
-
A database of anti-microbial peptides
- AMP:
-
Antimicrobial peptide
- BLAST:
-
Basic local alignment search tool
- BLP:
-
Bacteriocin like peptide
- CAMPR3:
-
Collection of anti-microbial peptides
- FA:
-
Formic acid
- FDR:
-
False discovery rate
- fmol:
-
Femtomole
- HPLC:
-
High performance liquid chromatography
- IPC:
-
Isoelectric point calculator
- kDa:
-
Kilodalton
- LAB:
-
Lactic acid bacteria
- LB:
-
Luria–Bertani
- LC–MS/MS:
-
Liquid chromatography–mass spectrometry and liquid chromatography–tandem mass spectrometry
- MALDI-TOF:
-
Matrix assisted laser desorption ionization-time of fight
- MASCOT:
-
Modular approach to software construction operation and test
- MGF:
-
Mascot generic format
- MRS:
-
Man, Rogosa, Sharpe
- MS:
-
Mass spectrometry
- MWCO:
-
Molecular weight cut-off
- NCBI:
-
National center for biotechnology information
- NRP:
-
Non-ribosomal peptide
- PBS:
-
Phosphate-buffered saline
- PNP:
-
Peptidic natural products
- RiPP:
-
Ribosomally synthesized and posttranslationally modified peptides
- TFA:
-
Trifluoroacetic acid
References
Montesinos E (2007) Antimicrobial peptides and plant disease control. FEMS Microbiol Lett 270(1):1–11. https://doi.org/10.1111/j.1574-6968.2007.00683.x
Mousa WK, Raizada MN (2015) Biodiversity of genes encoding anti-microbial traits within plant associated microbes. Front Plant Sci 6:25. https://doi.org/10.3389/fpls.2015.00231
Wang GS, Mishra B, Lau K, Lushnikova T, Golla R, Wang XQ (2015) Antimicrobial peptides in 2014. Pharmaceuticals 8(1):123–150. https://doi.org/10.3390/ph8010123
Hegedus N, Marx F (2013) Antifungal proteins: more than antimicrobials? Fungal Biol Rev 26(4):132–145. https://doi.org/10.1016/j.fbr.2012.07.002
van der Weerden NL, Bleackley MR, Anderson MA (2013) Properties and mechanisms of action of naturally occurring antifungal peptides. Cell Mol Life Sci 70(19):3545–3570. https://doi.org/10.1007/s00018-013-1260-1
Farkas A, Maroti G, Kereszt A, Kondorosi E (2017) Comparative analysis of the bacterial membrane disruption effect of two natural plant antimicrobial peptides. Front Microbiol 8:51. https://doi.org/10.3389/fmicb.2017.00051
Scocchi M, Mardirossian M, Runti G, Benincasa M (2016) Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem 16(1):76–88. https://doi.org/10.2174/1568026615666150703121009
Thwaite JE, Hibbs S, Titball RW, Atkins TP (2006) Proteolytic degradation of human antimicrobial peptide LL-37 by Bacillus anthracis may contribute to virulence. Antimicrob Agents Chemother 50(7):2316–2322. https://doi.org/10.1128/aac.01488-05
McGillivray SM, Ebrahimi CM, Fisher N, Sabet M, Zhang DX, Cheng YH, Haste NM, Aroian RV, Gallo RL, Guiney DG, Friedlander AM, Koehler TM, Nizet V (2009) ClpX Contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. Journal of Innate Immunity 1(5):494–506. https://doi.org/10.1159/000225955
Friedrich C, Scott MG, Karunaratne N, Yan H, Hancock REW (1999) Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob Agents Chemother 43(7):1542–1548. https://doi.org/10.1128/aac.43.7.1542
Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, Staubitz P, Adler-Moore J, McNamara PJ, Proctor RA, Yeaman MR, Bayer AS (2007) In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153:1187–1197. https://doi.org/10.1099/mic.0.2006/003111-0
Hincapié O, Giraldo P, Orduz S (2018) In silico design of polycationic antimicrobial peptides active against Pseudomonas aeruginosa and Staphylococcus aureus. Antonie Van Leeuwenhoek 111(10):1871–1882. https://doi.org/10.1007/s10482-018-1080-2
Porto WF, Irazazabal L, Alves ESF, Ribeiro SM, Matos CO, Pires Á, Fensterseifer ICM, Miranda VJ, Haney EF, Humblot V, Torres MDT, Hancock REW, Liao LM, Ladram A, Lu TK, de la Fuente-Nunez C, Franco OL (2018) In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat Commun 9(1):1490. https://doi.org/10.1038/s41467-018-03746-3
Liu S, Bao J, Lao X, Zheng H (2018) Novel 3D structure based model for activity prediction and design of antimicrobial peptides. Sci Rep 8(1):11189. https://doi.org/10.1038/s41598-018-29566-5
Piard JC, Desmazeaud M (1991) Inhibiting factors produced by lactic-acid bacteria. 1. Oxygen metabolites and catabolism end-products. Lait 71(5):525–541
Özogul F, Hamed I (2018) The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: a review. Crit Rev Food Sci Nutr 58(10):1660–1670. https://doi.org/10.1080/10408398.2016.1277972
Moshood Y, Tengku A (2013) Lactic acid bacteria: bacteriocin producer: a mini review. IOSR J Pharm 3(4):44–55
Gavrilova E, Anisimova E, Gabdelkhadieva A, Nikitina E, Vafina A, Yarullina D, Bogachev M, Kayumov A (2019) Newly isolated lactic acid bacteria from silage targeting biofilms of foodborne pathogens during milk fermentation. BMC Microbiol 19:248
Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CGM (2007) Bacteriocin production as a mechanism for the antfinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA 104(18):7617–7621. https://doi.org/10.1073/pnas.0700440104
Zamfir M, Callewaert R, Cornea PC, Savu L, Vatafu I, De Vuyst L (1999) Purification and characterization of a bacteriocin produced by Lactobacillus acidolphilus IBB 801. J Appl Microbiol 87(6):923–931. https://doi.org/10.1046/j.1365-2672.1999.00950.x
Messaoudi S, Manai M, Kergourlay G, Prevost H, Connil N, Chobert JM, Dousset X (2013) Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol 36(2):296–304. https://doi.org/10.1016/j.fm.2013.05.010
Giacometti A, Cirioni O, Ancarani F, Del Prete MS, Fortuna M, Scalise G (1999) In vitro activities of polycationic peptides alone and in combination with clinically used antimicrobial agents against Rhodococcus equi. Antimicrob Agents Chemother 43(8):2093–2096. https://doi.org/10.1128/aac.43.8.2093
Vaara M, Porro M (1996) Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob Agents Chemother 40(8):1801–1805. https://doi.org/10.1128/aac.40.8.1801
Yan TR, Lee CS (1997) Characterization of a partially purified bacteriocin, Fermentcin B, from Lactobacillus fermentum. Biotechnol Lett 19(8):741–744. https://doi.org/10.1023/a:1018327907435
Kaur B, Balgir PP, Mittu B, Kumar B, Garg N (2013) Biomedical Applications of fermenticin HV6b isolated from Lactobacillus fermentum HV6b MTCC10770. Biomed Res Int. https://doi.org/10.1155/2013/168438
Lima ET, Andreatti RL, Okamoto AS, Noujaim JC, Barros MR, Crocci AJ (2007) Evaluation in vitro of the antagonistic substances produced by Lactobacillus spp. isolated from chickens. Can J Vet Res 71(2):103–107
Dallas DC, Guerrero A, Parker EA, Robinson RC, Gan JN, German JB, Barile D, Lebrilla CB (2015) Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics 15(5–6):1026–1038. https://doi.org/10.1002/pmic.201400310
Bogachev MI, Kayumov AR, Markelov OA, Bunde A (2016) Statistical prediction of protein structural, localization and functional properties by the analysis of its fragment mass distributions after proteolytic cleavage. Sci Rep 6:19. https://doi.org/10.1038/srep22286
Yi LH, Luo LL, Lu X (2018) Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Front Microbiol 9:14. https://doi.org/10.3389/fmicb.2018.01567
Grafskaia EN, Polina NF, Babenko VV, Kharlampieva DD, Bobrovsky PA, Manuvera VA, Farafonova TE, Anikanov NA, Lazarev VN (2018) Discovery of novel antimicrobial peptides: a transcriptomic study of the sea anemone Cnidopus japonicus. J Bioinform Comput Biol 16(2):17. https://doi.org/10.1142/s0219720018400061
Khazigaleeva RA, Vinogradova SV, Petrova VL, Fesenko IA, Arapidi GP, Kamionskaya AM, Govorun VM, Ivanov VT (2017) Antimicrobial activity of endogenous peptides of the moss Physcomitrella patens. Russ J Bioorg Chem 43(3):248–254. https://doi.org/10.1134/s1068162017030062
Schrader M, Schulz-Knappe P, Fricker LD (2014) Historical perspective of peptidomics. EuPA Open Proteomics 3:171–182
Waghu FH, Barai RS, Gurung P, Idicula-Thomas S (2016) CAMP(R3): a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res 44(D1):D1094–D1097. https://doi.org/10.1093/nar/gkv1051
Lee HT, Lee CC, Yang JR, Lai JZC, Chang KY (2015) A large-scale structural classification of antimicrobial peptides. Biomed Res Int. https://doi.org/10.1155/2015/475062
Walsh CT (2004) Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303(5665):1805–1810. https://doi.org/10.1126/science.1094318
Mohimani H, Pevzner PA (2016) Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks. Nat Prod Rep 33(1):73–86. https://doi.org/10.1039/c5np00050e
Mohimani H, Liu WT, Yang YL, Gaudencio SP, Fenical W, Dorrestein PC, Pevzner PA (2011) multiplex de novo sequencing of peptide antibiotics. J Comput Biol 18(11):1371–1381. https://doi.org/10.1089/cmb.2011.0158
Allmer J (2011) Algorithms for the de novo sequencing of peptides from tandem mass spectra. Expert Rev Proteomics 8(5):645–657. https://doi.org/10.1586/epr.11.54
Novak J, Lemr K, Schug KA, Havlicek V (2015) CycloBranch: de novo sequencing of nonribosomal peptides from accurate product ion mass spectra. J Am Soc Mass Spectrom 26(10):1780–1786. https://doi.org/10.1007/s13361-015-1211-1
Fomin E (2016) A simple approach to the reconstruction of a set of points from the multiset of n(2) pairwise distances in n(2) steps for the sequencing problem: I. Theory. J Comput Biol 23(9):769–775. https://doi.org/10.1089/cmb.2016.0044
Fomin E (2016) A simple approach to the reconstruction of a set of points from the multiset of n(2) pairwise distances in n(2) steps for the sequencing problem: II. Algorithm. J Comput Biol 23(12):934–942. https://doi.org/10.1089/cmb.2016.0046
Mohimani H, Gurevich A, Mikheenko A, Garg N, Nothias LF, Ninomiya A, Takada K, Dorrestein PC, Pevzner PA (2017) Dereplication of peptidic natural products through database search of mass spectra. Nat Chem Biol 13(1):30–37. https://doi.org/10.1038/nchembio.2219
Tsapieva A, Duplik N, Suvorov A (2011) Structure of plantaricin locus of Lactobacillus plantarum 8P-A3. Benef Microbes 2(4):255–261. https://doi.org/10.3920/bm2011.0030
Schillinger U, Lücke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55(8):1901–1906
Stern NJ, Svetoch EA, Eruslanov BV, Perelygin VV, Mitsevich EV, Mitsevich IP, Pokhilenko VD, Levchuk VP, Svetoch OE, Seal BS (2006) Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob Agents Chemother 50(9):3111–3116. https://doi.org/10.1128/aac.00259-06
Zheng G, Slavik MF (1999) Isolation, partial purification and characterization of a bacteriocin produced by a newly isolated Bacillus subtilis strain. Lett Appl Microbiol 28(5):363–367. https://doi.org/10.1046/j.1365-2672.1999.00545.x
Klose V, Bayer K, Bruckbeck R, Schatzmayr G, Loibner AP (2010) In vitro antagonistic activities of animal intestinal strains against swine-associated pathogens. Vet Microbiol 144(3–4):515–521. https://doi.org/10.1016/j.vetmic.2010.02.025
Schulthess B, Bloemberg GV, Zbinden R, Bottger EC, Hombach M (2014) Evaluation of the Bruker MALDI biotyper for identification of gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J Clin Microbiol 52(4):1089–1097. https://doi.org/10.1128/jcm.02399-13
McGarvey JA, Franco RB, Palumbo JD, Hnasko R, Stanker L, Mitloehner FM (2013) Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J Appl Microbiol 114(6):1661–1670. https://doi.org/10.1111/jam.12179
Anisimova E, Yarullina D (2018) Characterization of erythromycin and tetracycline resistance in Lactobacillus fermentum strains. Int J Microbiol 2018:3912326–3912326. https://doi.org/10.1155/2018/3912326
Lane DJ (1991) 16S/23S rRNA sequencing. In: Goodfellow ESAM (ed) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Kozlowski LP (2016) IPC: isoelectric point calculator. Biol Direct 11:16. https://doi.org/10.1186/s13062-016-0159-9
Torrent M, Di Tommaso P, Pulido D, Nogues MV, Notredame C, Boix E, Andreu D (2012) AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics 28(1):130–131. https://doi.org/10.1093/bioinformatics/btr604
Gurevich A, Mikheenko A, Shlemov A, Korobeynikov A, Mohimani H, Pevzner PA (2018) Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra. Nat Microbiol 3(3):319–327. https://doi.org/10.1038/s41564-017-0094-2
Fomin E (2019) A simple approach to the reconstruction of a set of points from the multiset of pairwise distances in n2 steps for the sequencing problem: III. Noise inputs for the beltway case. J Comput Biol 26(1):68–75. https://doi.org/10.1089/cmb.2018.0078
Wilson DA, Young S, Timm K, Novak-Weekley S, Marlowe EM, Madisen N, Lillie JL, Ledeboer NA, Smith R, Hyke J, Griego-Fullbright C, Jim P, Granato PA, Faron ML, Cumpio J, Buchan BW, Procop GW (2017) Multicenter evaluation of the Bruker MALDI biotyper CA system for the identification of clinically important bacteria and yeasts. Am J Clin Pathol 147(6):623–631. https://doi.org/10.1093/ajcp/aqw225
Chanos P, Mygind T (2016) Co-culture-inducible bacteriocin production in lactic acid bacteria. Appl Microbiol Biotechnol 100(10):4297–4308. https://doi.org/10.1007/s00253-016-7486-8
Maldonado-Barragan A, Caballero-Guerrero B, Lucena-Padros H, Ruiz-Barba JL (2013) Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol 33(1):40–47. https://doi.org/10.1016/j.fm.2012.08.009
Guillot A, Boulay M, Chambellon E, Gitton C, Monnet V, Juillard V (2016) Mass spectrometry analysis of the extracellular peptidome of Lactococcus lactis: lines of evidence for the coexistence of extracellular protein hydrolysis and intracellular peptide excretion. J Proteome Res 15(9):3214–3224. https://doi.org/10.1021/acs.jproteome.6b00424
Hsu C, Wiseman GM (1972) The nature of epidermidins, new antibiotics from staphylococci. Can J Microbiol 18(2):121–125. https://doi.org/10.1139/m72-021
Funding
This study was funded by the Russian Foundation for Basic Research (Grant Nos. 17-00-00470 (K), 17-00-00456, 17-00-00461, 17-00-00462).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pavlova, A.S., Ozhegov, G.D., Arapidi, G.P. et al. Identification of Antimicrobial Peptides from Novel Lactobacillus fermentum Strain. Protein J 39, 73–84 (2020). https://doi.org/10.1007/s10930-019-09879-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-019-09879-8