Abstract
Breast carcinoma(BC)is the most common cancer type among females globally. Understanding the molecular pathways that trigger the development of BC is crucial for both prevention and treatment. As such, the role of transcription factors (TFs) in the development of BC is a focal point in this field. CREB3s play a critical role in initiating the unfolded protein response (UPR); however, the role of CREB3 family members in breast cancer development remains largely unknown. Here, we mined the ONCOMINE database for the transcriptional data of CREB3s in patients with BC. Then, the regulatory functions of a novel TF, CREB3L4, were investigated. CREB3L4 knockdown in MDA-MB-231 and MCF-7 cells suppressed proliferation and promoted apoptosis and cell cycle arrest. ChIP assays confirmed that CREB3L4 can directly bind to the PCNA promoter region, suggesting that the PCNA protein may be functionally downstream of CREB3L4. Additionally, the expression level of CREB3L4 was assessed using our cohort. CREB3L4 is upregulated in breast cancer tissues and is significantly associated with histological grade and tumour size (P = 0.001 and P < 0.001, respectively). Furthermore, PCNA expression was upregulated in breast cancer tissues and positively correlated with CREB3L4. In summary, CREB3L4 may play an important role in the progression of human BC and may serve as a therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the latest GLOBOCAN statistics, breast carcinoma (BC) is the most common invasive cancer in women and the second leading cause of cancer-related death worldwide [1]. The latest hypothesis is that appropriately inhibiting oncogenic transcription factors (TFs) is a target for BC treatment [2]. TFs are often the ultimate regulators of several central processes, such as proliferation, survival, self-renewal and invasion. Under different pathological conditions, TFs may aberrantly initiate the transcription of cancer-related genes, which increases cell proliferation, differentiation, and angiogenesis and suppresses apoptosis or cell-cell adhesions, all of which consequently lead to cancer development [3].
Cyclic adenosine monophosphate (cAMP) response element binding (CREB) [4], a phosphorylation-dependent TF, is overexpressed and constitutively phosphorylated in numerous human cancers [5] and plays a critical role in disease pathogenesis and prognosis [6] and cancer cell migration [7]. CREB belongs to the basic leucine zipper (bZIP)-containing TF superfamily, which includes mammalian C/EBP, c-Jun, c-Fos and c-Myc [8]. Among these bZIP factors, CREBs, activating transcription factors(ATFs), and cAMP response element modulator (CREM) constitute a subgroup named ATF/CREB based on the extent of their homology at the bZIP region [9]. Among the members of this subgroup, the CREB3 proteins and ATF6 are substrates for S1P/S2P [10]. These proteins possess a transmembrane segment, localize to the endoplasmic reticulum (ER) and are activated by regulated intramembranous proteolysis, suggesting that they could play a critical role in initiating the unfolded protein response (UPR) [11], which is widely considered a mechanism of tumour progression [12].
The distinctive CREB3 subfamily consists of five members [11, 13]: CREB3 (LZIP/Luman) [14], CREB3L1 (OASIS) [15], CREB3L2 (BBF2H7) [16], CREB3L3 (CREB-H) [17] and CREB3L4 (Tisp40, AIbZIP, or ATCE1) [18]. CREB3L4, a novel human CREB gene, is localized on chromosome 1q21.3 and encodes a protein comprising 395 amino acid residues. As is typical of other ATF/CREB proteins, CREB3L4 contains a bZIP domain, which is located in its central region (amino acids 222–280). In humans, CREB3L4 is highly expressed in a variety of cancer types, including prostate carcinomas [11, 19, 20], hepatocellular carcinomas [21] and endometrial cancers [13]. Recently, it was revealed that the high messenger RNA (mRNA) expression of CREB3L4 was associated with reduced relapse-free survival (RFS) and disease-specific survival (DSS) in BC [22]. However, further studies investigating the mRNA expression of all CREB3 members in BC are necessary. In addition, the mechanism(s) responsible for CREB3L4 overexpression also need to be investigated, and it is unclear whether CREB3L4 overexpression is a consequence of or a contributing factor to cancer development.
The purpose of the current study was twofold. The first objective was to focus on the importance and relevance of the CREB3 family in BC and to identify which family member is most involved in the development of breast cancer. The second objective was to identify a possible target gene of CREB3L4. Data pertaining to all of these factors should considerably increase our understanding of the function of CREB3L4.
Materials and Methods
ONCOMINE Database Analysis
Gene expression array datasets from ONCOMINE (http://www.oncomine.org), a publicly online database containing 715 datasets and 86,733 samples [23], were searched to examine the transcription levels of the different CREB3s subfamily members in different cancers. The mRNA expression of these genes (log2-transformed) in clinical cancer specimens was compared with that in normal controls samples using Student’s t test to generate a P value. To identify the CREB3 proteins with the most significant expression, the following parameters were defined: P < 0.001, fold change >2 and gene ranking in the top 10%.
Cells and Tissues
Four human breast cancer cell lines, MDA-MB-231, MDA-MB-468, MCF-7 and T-47D, were cultured at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (HyClone, USA) under a 5% CO2 atmosphere. MCF10A cells were cultured in DMEM-F12 (Hyclone, China) supplemented with 10 μg/ml insulin (Gibco), 0.5 μg/ml hydrocortisone (Sigma), 15 ng/ml epidermal growth factor (Sigma), 5% horse serum (Gibco), and 100 ng/ml cholera toxin (Sigma) and incubated at 37 °C in a humidified incubator with 5% CO2. This study was approved by the Human Ethics Review Committee of Qilu Hospital of Shandong University (Qingdao). The use of 80 pairs of breast cancer tumour and matched adjacent normal tissues was approved by the Department of Pathology of Qilu Hospital of Shandong University (Qingdao). All patients underwent surgical resection at Qilu Hospital of Shandong University (Qingdao). Informed consent was obtained from all subjects or their relatives. All of the tissue samples were snap-frozen and stored at −80 °C in liquid nitrogen.
Lentivirus Infection
Lentiviruses carrying a short hairpin RNA (shRNA) target sequence for shCREB3L4 (5’-CGGAGTGACTTCCAGAAATAT-3′) were obtained from GeneChem. The viruses produced were used in the presence of polybrene to infect MCF7 and MDA-MB-231 cells. Forty-eight hours later, MCF7 and MDA-MB-231 cells were cultured in medium containing puromycin for the selection of clones with stable CREB3L4 knock down, which were identified and verified by Western blotting using the indicated primary antibodies.
Real-Time PCR
Total RNA was extracted using TRIzol reagent (TaKaRa) following the manufacturer’s protocol. The miScript Reverse Transcription Kit (TaKaRa) was used for quantitative real-time polymerase chain reaction (qRT-PCR). SYBR green PCR Master Mix (TaKaRa) was used for the detection of mRNA expression following the manufacturer’s protocol. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression was used as an endogenous control. The 2-ΔΔCT method was used to analyse the relative fold changes in expression. The primers were designed by Sangon Biotech (Shanghai, China). Their sequences are as follows: CREB3L4, forward: 5’-CAGACGCTAATTGCTCAAACTTC-3′ and reverse 5’-CCACTTGGGTCTCCAGATTTTCT-3′; PCNA, forward 5’-CCTGCTGGGATATTAGCTCCA-3′ and reverse 5’-CAGCGGTAGGTGTCGAAGC-3′; and GAPDH, forward 5’-GGAGCGAGATCCCTCCAAAAT-3′ and reverse 5’-GGCTGTTGTCATACTTCTCATGG-3′.
Immunoblot Assay
Cells were lysed with RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA), and the resulting protein samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) through a 10% gel and transferred onto PVDF membranes (Millipore). The membranes were blocked with 5% nonfat milk at room temperature for 2 h before they were incubated overnight with primary antibodies at 4 °C. After they were washed with TBST three times for 15 min each, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature with gentle shaking. GAPDH was used as the loading control. Immunoreactive bands were visualized using SuperSignal West Pico Chemiluminescent Substrates (Thermo, USA). The primary antibodies used in this study were anti-CREB3L4 (Thermo Fisher, PA5–18028), anti-PCNA (Thermo Fisher, MA5–11358), and anti-GAPDH (Abcam, ab181602).
Proliferation Assay
Cells were assessed for proliferation by measuring 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt Sigma (MTS) reduction. Cells were seeded into 96-well plates (100 μl cell suspensions, 1 × 103 cells/ml). Twenty-four hours later, 0.05 mg ml−1 MTS reagent (Promega) was added to each well and incubated at 37 °C for 4 h followed by measurement of the absorbance at 490 nm. The values were standardized to those of wells containing media alone.
Apoptosis Assay
MCF7 and MDA-MB-231 cells were transfected with shCtrl and shCREB3L4. Forty-eight hours later, transfected cells were treated with cisplatin (10 μM, 24 h, Sigma). The apoptotic cells were then washed with phosphate-buffered saline (PBS) and stained with fluorescein isothiocyanate-annexin V and propidium iodide (PI) according to the manufacturer’s protocol (Beijing Biosea Biotechnology Annexin V Kit). Apoptotic cells (annexin V-positive, PI-negative) were then determined by flow cytometry.
Cell Cycle Assay
Cell cycle analysis was performed by monitoring the DNA content at various time points with PI staining. Cells synchronized at S phase with a double thymidine (Sigma, 2 mM) block were released, cultured in fresh medium for 12 h, fixed in 70% cold ethanol, incubated on ice for 2 h, washed once in PBS containing 0.1% glucose, and incubated for 30 min at 37 °C in PBS containing 0.1% glucose, 1 mg of RNase A (Thermo) per ml, and 20 μg of PI per ml. DNA content was determined by flow cytometry.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using an iDeal ChIP-seq Kit for Transcription Factors (Diagenode, C01010170) with antibodies against CREB3L4 and PCNA. A lysate of 5 × 106 cells was used for each ChIP reaction. Precipitated DNA samples were analysed by quantitative PCR with primer pairs specific to the promoters of the PCNA and CREB3L4 genes. qRT-PCR was used to quantify gene expression as previously described. The ChIP primers used were as follows: forward, 5’-ATGGACACGATTGGCCCTAAA-3′; reverse, 5’-CTACGCCTGCAACCGTTTAAT-3′.
Luciferase Reporter Assay
PCNA promoter cloning: The region of the PCNA promoter was cloned by PCR from normal human breast cDNA. Primers were listed in the followed table. The sequenced fragments were inserted into the to the promoter less plasmid pGL2-enhancer control vector using SacI and NarI enzyme.
PCR | Primers | Sequence (5′-3′) |
|---|---|---|
Promoter cloning | Upstream | ATTATGTTTTGATATTGAGATGTATGCAA |
Downstream | CAAGCGCGCGCTCTCACCCTGCGCCGCGT |
Immunohistochemical (IHC) Staining
Haematoxylin and eosin (H&E) staining was performed to differentiate cancer and normal breast tissues in the same section. Immunohistochemistry was performed by using the avidin-biotin complex method (ABC; Vector Laboratories, Burlingame, CA), which included heat-induced antigen retrieval procedures. Incubation with antibodies against CREB3L4 (PA5–58254, 1:500 dilution, Thermo Fisher) and PCNA (MA5–11358, 1:400 dilution, Thermo Fisher) was carried out at 4 °C for 18 h. All staining was assessed by pathologists blinded to the diagnosis of the subjects and their outcomes. The widely accepted German semiquantitative scoring system that considers the staining intensity and extent of staining was used. Each specimen was assigned a score according to the intensity of the nucleic, cytoplasmic, and/or membrane staining (no staining = 0; weak staining = 1, moderate staining = 2, strong staining = 3) and the percentage of stained cells (0% = 0, 1–24% = 1, 25–49% = 2, 50–74% = 3, 75–100% = 4). The final immunoreactive score was determined by multiplying the intensity score by the percentage score, which ranged from 0 (the minimum score) to 12 (the maximum score). A score ≥ 5 was considered positive staining, and a score ≤ 4 was considered negative staining.
Statistical Analysis
The immunohistochemistry results were analysed by the χ2 test, and all other results are expressed as the mean ± standard deviation (SD) derived from three independent experiments unless stated otherwise. Student’s t test was used for statistical comparisons between means where applicable. Differences among more than two groups in the above assays were estimated using analysis of variance (ANOVA). A correlation analysis was performed with Spearman’s rank correlation coefficient. All statistical analyses were performed with GraphPad Prism 5 and SPSS 18.0 software. All statistical tests were two-sided, and P values <0.05 were considered statistically significant.
Results
The Transcription Levels of CREB3 Subfamily Members in BC
As mentioned before, five CREB3 subfamily members have been identified [13]. First, we analysed the differences in the transcription levels of these genes between tumour and normal tissues in multiple types of cancer obtained from the online ONCOMINE database. As shown in Fig. 1, the ONCOMINE database contained a total of 408, 412, 346, 251 and 278 unique analyses for CREB3, CREB3L1, CREB3L2, CREB3L3 and CREB3L4, respectively. CREB3L4 was observed in 4 human cancer types, including haematological malignancies and solid tumours. The mRNA expression level of CREB3L4 was significantly increased in BC samples in 8 studies. In The Cancer Genome Atlas (TCGA) dataset, CREB3L4 overexpression was observed in all types of BC compared with normal tissue, including invasive ductal and lobular BC (fold change = 4.927), mixed lobular and ductal BC (fold change = 4.663), invasive lobular BC (fold change = 3.379), and invasive ductal BC (fold change = 2.594). In the Gluck [24] dataset, CREB3L4overexpression was observed in invasive BC compared to normal tissue (fold change =3.391). In the Curtis [25] dataset, CREB3L4 overexpression was observed in tubular BC (fold change = 2.638), mucinous BC (fold change = 2.162), and invasive ductal BC (fold change = 2.045) compared to the respective normal tissues (Table 1).
The mRNA expression levels of CREB3s subfamily members in ONCOMINE datasets of different types of human cancers were analysed. These results highlight the numbers of datasets with statistically significant (p < 0.01) mRNA overexpression (red) or underexpression (blue) of the target gene (cancer vs. normal tissue). Cell colour was determined by the best gene rank percentile for the analyses within the cell. The gene rank was analysed by the percentile of target genes in the top of all genes measured in each experiment. The p value threshold was 0.001, and the fold change was defined as 2
Another gene showing increased mRNA expression according to the 4 datasets was CREB3L1. In the TCGA dataset, CREB3L1 overexpression was found in invasive ductal and lobular BC (fold change = 2.524), mixed lobular and ductal BC (fold change = 1.606), invasive lobular BC (fold change = 2.837), and invasive ductal BC (fold change = 2.162). In the Gluck dataset, CREB3L1 overexpression was observed in invasive BC compared to normal tissues (fold change =2.135) (Table 1). However, neither CREB3 nor CREB3L2 mRNA was detected in breast cancer tissue samples. In addition, the mRNA expression of CREB3L3 was decreased in BC according to 3 datasets (Fig. 1). These results suggest that CREB3L4 may be an oncogenic factor and play a specific role in the occurrence and development of BC.
CREB3L4 Is Upregulated in Breast Cancer Cells
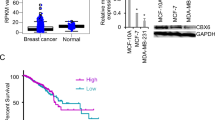
On the basis of Northern blot analyses, full-length CREB3L4 mRNA is expressed only in the human prostate as well as in breast and prostate cancer cell lines [19]. Therefore, RT-qPCR and Western blot analyses were performed to detect CREB3L4 expression in BC cells. As shown in Fig. 2a, CREB3L4 was upregulated not only in oestrogen receptor-positive MCF-7 and T-47D breast cancer cells but also in the triple-negative breast cancer (TNBC) cell lines MDA-MB-231 and MDA-MB-468, especially in the noninvasive MCF7 and highly aggressive MDA-MB-231 cells. To investigate the function of CREB3L4 in BC cell lines, MDA-MB231 and MCF7 cells with stable CREB3L4 knockdown were established by the introduction of a shRNA expression vector. As confirmed by real-time PCR and Western blot, the results showed that endogenous CREB3L4 expression was more downregulated in cells transduced with shCREB3L4 than in cells transduced with the control (Fig. 2b, c).
CREB3L4 is upregulated in breast cancer cells. (a) Analysis of CREB3L4 mRNA and protein expression in the BC cell lines MDA-MB-231, MDA-MB-468, MCF-7 and T-47D by RT-PCR and western blot. The highest CREB3L4 expression among the breast cancer cell lines was observed in MCF-7 and MDA-MB-231 cells (p < 0.05). (b) The mRNA level of CREB3L4 in MCF-7 (p < 0.01) and MDA-MB-231 (p < 0.05) cells transfected with CREB3L4 shRNA was significantly reduced compared to that in nontransfected cells. (c) The protein expression of CREB3L4 in MCF-7 and MDA-MB-231 cells transfected with CREB3L4 shRNA was detected with immunoblotting. The data are presented as the means ± SDs of three independent experiments. *p < 0.05 and **p < 0.01. PCR: polymerase chain reaction; IB: immunoblot
Downregulation of CREB3L4 Suppresses Proliferation and Induces Cell Cycle Arrest and Apoptosis in BC Cells
Next, we analysed the effect of CREB3L4 on breast cancer cells. We applied the MTS assay to evaluate cell proliferation. Three days after transfection, a reduction in cell viability was observed in the shCREB3L4 group, and the inhibitory effect became more significant at 4 and 5 days after transfection (P < 0.01). Knockdown of CREB3L4 in MCF7 and MDA-MB-231 cells significantly decreased the extent of cell proliferation compared to that of the respective negative control cells (Fig. 3a). This result suggests that CREB3L4 promotes the proliferation of BC cells. In addition, annexin V/PI double-staining and PI staining tests were used to evaluate apoptosis and cell cycle distribution. The results revealed that cells with CREB3L4 knockdown were significantly more sensitive to cisplatin-induced apoptosis than were the negative control cells (P < 0.05) (Fig. 3b). Moreover, we analysed cell cycle distribution using flow cytometry, which showed that CREB3L4 knockdown led to cell cycle arrest at S phase in both MCF-7 and MDA-MB-231 cells at 12 h after release from the thymidine block (P < 0.01, Fig. 3c). These results indicate that CREB3L4 downregulation greatly promoted apoptosis and cell cycle arrest in both MCF-7 and MDA-MB-231 cells. In conclusion, we demonstrated that CREB3L4 could modulate the cellular function of BC cells and may play an oncogenic role in breast cancer.
The function of CREB3L4 in BC cell proliferation, apoptosis and cell cycle arrest. (a) The proliferation activities of MCF-7 and MDA-MB-231 cells transfected with shCtrl and shCREB3L4 were determined by the MTS assay. (b) The percentages of apoptotic cells in MCF-7 and MDA-MB-231 cells transfected with shCtrl and shCREB3L4 were evaluated by flow cytometry. (c) The percentages of PI-stained cells at G1, S and G2/M phases in MCF-7 and MDA-MB-231 cells transfected with shCtrl and shCREB3L4 were determined by flow cytometry. The data are presented as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01
CREB3L4 Binds to the Promoter Region of PCNA
Based on the understanding of the pathologic role of CREB3L4, we continued to explore the underlying regulatory mechanisms. Thus, we investigated the promoter region of PCNA and found that the sequence “GTGACGTC” was located in its promoter region (Fig. 4a). The results of the ChIP assay indicated that CREB3L4 directly binds to the PCNA promoter region, and both MCF7 and MDA-MB-231 cells with CREB3L4 silencing exhibited reduced CREB3L4 binding with the PCNA promoter (Fig. 4b).This binding can alter gene transcription was also tested by luciferase promoter assay (Fig. 4c). Next, we found that silencing CREB3L4 reduced the protein level of PCNA (Fig. 4d), suggesting a regulatory effect of CREB3L4 on PCNA.
CREB3L4 binds to the promoter region of PCNA. (a) CREB3L4 binding sites present in the PCNA promoter region. (b) ChIP assays were performed to examine CREB3L4-bound DNA using anti-IgG. ChIP was carried out with anti-IgG (negative control) and anti-CREB3L4 antibodies in MCF7 and MDA-MB-231 cells. The qPCR results for the PCNA promoter region are shown as the percentage of input DNA. (c) The promoter-less pGL2 control vector and PCNA promoter vector was transected with CREB3L4 plasmid into MCF7 and MDA-MB-231 cells, and the activity of the region was tested by dual-luciferase reporter assay. The data was shown as mean ± SEM for three independent experiments. (d)The protein expression levels were detected by Western blotting analysis using GAPDH as an internal control. Data are shown as the means ± SDs. ** p < 0.01
Expression and Correlation of CREB3L4 and PCNA in Breast Cancer Tissues
The relationships between CREB3L4 expression and the clinicopathological features of breast cancers are summarized in Table 2. CREB3L4 is up-regulated in breast cancer tissues and is significantly associated with histological grade and tumour size (P = 0.001 and P < 0.001, respectively). However, the associations between CREB3L4 expression and patient age, oestrogen receptor (ER), progesterone receptor (PR), HER2, and lymph node status were not statistically significant. The TCGA data set was also used to analyse the correlation between CREB3L4 and ER/PR/HER2, which are typical biomarkers of breast cancer. However, we still found no correlation between CREB3L4 expression and ER/PR/HER2 (R2 = 0.1849/0.1024/0.1849, Fig. 5a). CREB3L4 expression was upregulated in tissue samples derived from patients with a high histologic grade and a large tumour size (Fig. 5b).
The expression and correlation of CREB3L4 and PCNA in BC and nontumour tissues. (a) The correlation between CREB3L4 and ER/PR/HER2 was analysed with the TCGA data set. The data is presented as Log fold expression. (b) Representative images from the immunohistochemical staining of CREB3L4 in three serial sections of the same tumour and matched adjacent normal tissues. Scale bars, 50 μm. CREB3L4 protein expression in BC tissues is shown according to histological grade and tumour size. (magnification, ×400; p < 0.05). (c) CREB3L4 and PCNA mRNA expression levels in 32 paired BC and nontumour tissues were examined using real-time PCR. (d) Positive correlation of CREB3L4 with PCNA protein levels. A statistically significant association between CREB3L4 and PCNA expression was observed in breast cancer specimens (n = 80). **p < 0.01
Since the molecular mechanism of the interaction between CREB3L4 and PCNA was verified, we further analysed the correlation between CREB3L4 and PCNA expression in tumour tissues using clinical breast cancer samples. The mRNA expression of CREB3L4 and PCNA in 32 pairs of BC tissues and adjacent nontumour tissues was examined using real-time PCR. The levels of CREB3L4 (P = 0.0108) and PCNA (P = 0.001) mRNA expression were significantly higher in cancerous tissues than in normal tissues (Fig. 5c). To assess the consistency of protein translation with the observed transcription levels, the protein levels of CREB3L4 and PCNA in 80 pairs of BC tissues and adjacent nontumour tissues were examined. The analysis of the relationship between CREB3L4 expression and PCNA expression showed a significantly positive correlation (R = 0.9139, 95% CI: 0.8686–0.9441, p < 0.001) (Fig. 5d). These results suggest that CREB3L4 is upregulated in breast cancers concomitant with the increase in PCNA expression to promote tumorigenesis. Altogether, our clinical results indicate that CREB3L4 may play a role in the tumorigenesis of BC.
Discussion
CREB typically serves as a proto-oncogenic TF, and its role in human cancers is tissue- and/or cell type-specific [5]. CREB is elevated in breast cancer tissues [26] and positively regulates the proliferation and metastasis of breast cancer as well as bone destruction in breast cancer patients [27]. Among the CREB proteins, CREB3 subfamily members are particularly involved in the UPR. In breast tumours, chronic activation of the UPR is associated with therapy resistance and disease recurrence [28, 29]. However, studies on CREB3s in BC are rare. The development of microarray technology has become an essential component of biology and biomedical research [30]. Therefore, we performed an analysis of the expression of different CREB3 subfamily members in BC patients based on thousands of online analyses on gene expression or copy number variation. These results suggest that CREB3L1 and CREB3L4 mRNA levels are increased in breast cancer tissues. Alison K. Ward demonstrated that decreased CREB3L1 mRNA expression was associated with increased tumour grade and reduced progression-free survival. CREB3L1 is frequently upregulated in luminal and HER2-amplified breast cancer but not in TNBC [31]. However, recently, Bray Denard1 reported that TNBC tumours with high CREB3L1 expression have a high response rate to doxorubicin-based chemotherapy [32]. Through a comparative analysis, we found that CREB3L4 is a member of the CREB3 subfamily and is more closely related to the development of breast cancer than is CREB3L1. Further analysis of the clinical samples showed that CREB3L4 is highly expressed in BC tissues and positively correlated with tumour size and histological grade. However, the TCGA dataset and our cohort data indicate that CREB3L4 is independent of molecular typing. Our current clinical sample follow-up time is too short to provide meaningful clinical outcome data, and we feel it is too preliminary to make any conclusions at this stage.
We also elucidated its roles in the proliferation, apoptosis and cell cycle arrest of breast cancer cells. First, we generated shRNA to assess the potential role of CREB3L4 in cancer cell proliferation. The results showed that CREB3L4 knockdown significantly reduced cell viability and inhibited breast cancer proliferation. Furthermore, CREB3L4 knockdown also resulted in apoptosis and cell cycle arrest of breast cancer cells. Thus, CREB3L4 is a newly discovered TF associated with the development of breast cancer.
Oestrogen plays an important role in the development and physiology of the normal breast as well as in breast cancer cell proliferation. CREB3L4 has been reported to be highly expressed in sex-related cancers, including prostate, endometrium and uterine cancers [13]. In our experiment, CREB3L4 expression was upregulated in both oestrogen receptor-positive and TNBC cell lines. However, we did not detect a correlation between CREB3L4 protein expression and ER/PR/Her2 expression. In addition, CREB has emerged as a novel effector of p21-activated kinase 4 (PAK4); however, the upstream regulators in the ER/AR and WNT signalling pathways remain to be determined [33]. We wanted to determine whether CREB3L4 plays an important role in oestrogen-induced breast development and oestrogen-dependent breast cancer and whether CREB3L4 is downstream of PAK4. However, to confirm these hypotheses, we will need to quantitate CREB3L4 expression in a larger number of tumour specimens that are collected and processed under standardized conditions. In future studies, it will be of interest to determine whether CREB3L4 expression correlates with other clinically relevant endpoints. These findings will determine whether CREB3L4 may serve as a good immunohistochemical marker for breast cancer cells.
As CREB is a phosphorylation-dependent TF, its phosphorylation is sufficient to induce cellular gene expression in response to mitogen or stress-related signals [34]. Phosphorylation at Ser133 is required for CREBs to bind to CREB-binding proteins (CBPs) [35]; however, PAK4-mediated CREB activation is independent of its phosphorylation at Ser133. Rather, PAK4 activates CREB by phosphorylating CREB-regulated transcriptional cofactor 1 (CRTC1), a coactivator of CREB [36]. The phosphorylation status and phosphorylation site of CREB3L4 need to be further studied in breast cancer. Additionally, whether this mechanism is involved in BC tumorigenesis remains to be ascertained.
CREB mediates the activation of cAMP-responsive genes by binding as a dimer to a conserved cAMP-responsive element (CRE), TGACGTCA [37, 38] .Mayr and Marc Montminy had predicted that PCNA is one of the target genes containing consensus sites for CREB binding [34]. PCNA is a kind of cycle-related protein that is an indicator of tumour proliferation and is related to the occurrence of early cancer and tumour progression [39, 40]. Its main function is to participate in DNA replication, and it is also involved in DNA excision repair cell cycle control chromatin assembly and RNA transcription [41]. PCNA interacts with the S-phase-specific cyclin-dependent kinase 2 (CDK2)-cyclin A complex to stimulate the phosphorylation of CDK2 substrates, thereby targeting the CDK2-cyclin A complex to bind to DNA replication proteins [42] .ILK overexpression in breast cancer cells promoted proliferation and cell activity by activating the PI3K/Akt pathway and upregulating PCNA [43].To further investigate the regulatory mechanism by which CREB3L4 plays an oncogenic role, we determined its downstream target gene, PCNA. ChIP assays can realistically reflect the binding of TFs to the promoter in cells, and the results indicated that CREB3L4 directly binds the promoter of PCNA. This binding can alter gene transcription was also tested by luciferase promoter assay. IHC assays on clinical samples demonstrated that CREB3L4 and PCNA protein expression was positively correlated. These results suggest that CREB may promote PCNA expression by directly binding to the PCNA promoter, supporting the notion that a CREB3L4/PCNA axis exists in BC cells. Thus, their regulation and underlying molecular mechanism in BC cells need to be further studied. Whether a novel CREB3L4/PCNA axis participates in the biological behaviours of BC cell is still a key problem to be addressed in our future studies.
Conclusion
Taken together, our results indicate that, among the CREB3 subfamily members, CREB3L4 is most closely linked to the development of breast cancer. Our study demonstrated that CREB3L4 promotes BC oncogenesis via increased proliferation and reductions in apoptosis and cell cycle arrest in breast cancer cells. CREB3L4 binds to the PCNA promoter region. Both are highly expressed in breast cancer tissues, with a positive correlation. Thus, the CREB3L4/PCNA axis may play a crucial role in tumour progression. CREB may upregulate PCNA expression, leading to breast cancer cell proliferation and migration. Therefore, CREB3L4 may be a promising therapeutic target for breast cancer.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Hasanpourghadi M, Pandurangan AK, Mustafa MR. Modulation of oncogenic transcription factors by bioactive natural products in breast cancer. Pharmacol Res. 2018;128:376–88.
Hsieh WT, et al. Transcription factor and microRNA-regulated network motifs for cancer and signal transduction networks. BMC Syst Biol. 2015;9(Suppl 1):S5.
Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328(6126):175–8.
Wang YW, Chen X, Ma R, Gao P. Understanding the CREB1-miRNA feedback loop in human malignancies. Tumour Biol. 2016;37(7):8487–502.
Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15(8):2583–7.
Singh R, Shankar BS, Sainis KB. TGF-beta1-ROS-ATM-CREB signaling axis in macrophage mediated migration of human breast cancer MCF7 cells. Cell Signal. 2014;26(7):1604–15.
Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–6.
Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–48.
Stirling J, O'Hare P. CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol Biol Cell. 2006;17(1):413–26.
Labrie C, Lessard J, Ben Aicha S, Savard MP, Pelletier M, Fournier A, et al. Androgen-regulated transcription factor AIbZIP in prostate cancer. J Steroid Biochem Mol Biol. 2008;108(3–5):237–44.
Sisinni, L., et al., Endoplasmic Reticulum Stress and Unfolded Protein Response in Breast Cancer: The Balance between Apoptosis and Autophagy and Its Role in Drug Resistance. Int J Mol Sci, 2019. 20(4).
Cui X, et al. The androgen-induced protein AIbZIP facilitates proliferation of prostate cancer cells through downregulation of p21 expression. Sci Rep. 2016;6:37310.
Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17(9):5117–26.
Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun. 2002;293(1):470–7.
Storlazzi CT, Mertens F, Nascimento A, Isaksson M, Wejde J, Brosjo O, et al. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum Mol Genet. 2003;12(18):2349–58.
Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, et al. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29(10):2154–62.
Cao G, Ni X, Jiang M, Ma Y, Cheng H, Guo L, et al. Molecular cloning and characterization of a novel human cAMP response element-binding ( CREB) gene ( CREB4). J Hum Genet. 2002;47(7):373–6.
Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, et al. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 2002;62(3):721–33.
Kim TH, et al. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep. 2017;7:45300.
Inagaki Y, Yasui K, Endo M, Nakajima T, Zen K, Tsuji K, et al. CREB3L4, INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2008;180(1):30–6.
Jing X, Liang H, Hao C, Yang X, Cui X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol Rep. 2019;41(2):801–10.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6.
Gluck S, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/− trastuzumab. Breast Cancer Res Treat. 2012;132(3):781–91.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52.
Chhabra A, Fernando H, Watkins G, Mansel RE, Jiang WG. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep. 2007;18(4):953–8.
Son J, Lee JH, Kim HN, Ha H, Lee ZH. cAMP-response-element-binding protein positively regulates breast cancer metastasis and subsequent bone destruction. Biochem Biophys Res Commun. 2010;398(2):309–14.
Clarke R, Shajahan AN, Wang Y, Tyson JJ, Riggins RB, Weiner LM, et al. Endoplasmic reticulum stress, the unfolded protein response, and gene network modeling in antiestrogen resistant breast cancer. Horm Mol Biol Clin Investig. 2011;5(1):35–44.
Clarke R, Cook KL. Unfolding the role of Stress response signaling in endocrine resistant breast cancers. Front Oncol. 2015;5:140.
Rao A, P D, Renumadhavi CH, Chandra MG, Srinivasan R. Compressed sensing methods for DNA microarrays, RNA interference, and metagenomics. J Comput Biol. 2015;22(2):145–58.
Ward AK, Mellor P, Smith SE, Kendall S, Just NA, Vizeacoumar FS, et al. Epigenetic silencing of CREB3L1 by DNA methylation is associated with high-grade metastatic breast cancers with poor prognosis and is prevalent in triple negative breast cancers. Breast Cancer Res. 2016;18(1):12.
Denard B, Jiang S, Peng Y, Ye J. CREB3L1 as a potential biomarker predicting response of triple negative breast cancer to doxorubicin-based chemotherapy. BMC Cancer. 2018;18(1):813.
Won SY, Park JJ, Shin EY, Kim EG. PAK4 signaling in health and disease: defining the PAK4-CREB axis. Exp Mol Med. 2019;51(2):11.
Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609.
Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–61.
Won SY, et al. Nigral dopaminergic PAK4 prevents neurodegeneration in rat models of Parkinson's disease. Sci Transl Med. 2016;8(367):367ra170.
Klieman RL, Stephenson SH. Calcium antagonists--drug interactions. Rev Drug Metab Drug Interact. 1985;5(2–3):193–217.
Short JM, Wynshaw-Boris A, Short HP, Hanson RW. Characterization of the phosphoenolpyruvate carboxykinase (GTP) promoter-regulatory region. II. Identification of cAMP and glucocorticoid regulatory domains. J Biol Chem. 1986;261(21):9721–6.
Mao Y, Zhang Y, Fan S, Chen L, Tang L, Chen X, et al. GALNT6 promotes Tumorigenicity and metastasis of breast Cancer cell via beta-catenin/MUC1-C signaling pathway. Int J Biol Sci. 2019;15(1):169–82.
Tommasi S, Pfeifer GP. In vivo structure of two divergent promoters at the human PCNA locus. Synthesis of antisense RNA and S phase-dependent binding of E2F complexes in intron 1. J Biol Chem. 1999;274(39):27829–38.
Jurikova M, et al. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–52.
Koundrioukoff S, Jónsson ZO, Hasan S, de Jong RN, van der Vliet P, Hottiger MO, et al. A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol Chem. 2000;275(30):22882–7.
Qu Y, Hao C, Xu J, Cheng Z, Wang W, Liu H. ILK promotes cell proliferation in breast cancer cells by activating the PI3K/Akt pathway. Mol Med Rep. 2017;16(4):5036–42.
Acknowledgements
Qian Pu would like to give the greatest thanks to Haiyun Song and Qianqian Zhao of the Pathology Department of Qilu Hospital (Qingdao) for their help in this study.
Funding
This work was supported by Chinese National Natural Science Foundation Projects (Grant Nos. 81,572,587, 81,602,310).
Author information
Authors and Affiliations
Contributions
HDG conceived of and designed the experiments. QP and YRL designed and performed the experiments and wrote the paper. KD and LL performed the experiments. WWG analysed the data. HDG critically reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that there are no conflicts of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Research Ethics Committee of Qilu Hospital (Qingdao) of Shandong University and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pu, Q., Lu, L., Dong, K. et al. The Novel Transcription Factor CREB3L4 Contributes to the Progression of Human Breast Carcinoma. J Mammary Gland Biol Neoplasia 25, 37–50 (2020). https://doi.org/10.1007/s10911-020-09443-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-020-09443-6