Abstract
Background
Doxorubicin-based chemotherapy is currently the most frequently used treatment for triple negative breast cancer (TNBC), yet the response rate is not high due to the lack of a biomarker allowing identification of responsive patients before the chemotherapy is initiated. We have demonstrated that doxorubicin inhibits proliferation of cancer cells through proteolytic activation of a transcription factor called CREB3L1 (cAMP response element binding protein 3-like 1), and that CREB3L1 expression in cancer cells is a key determinant of their sensitivity to doxorubicin when they are cultured in vitro or established as xenograft tumors in mice. The purpose of this study is to determine whether CREB3L1 expression in tumor cells of TNBC patients can be established as a biomarker to predict outcomes of doxorubicin-based chemotherapy.
Methods
We performed a retrospective analysis on breast core biopsy tissue samples taken from 18 TNBC patients before they were treated with doxorubicin-based chemotherapy. CREB3L1 expression in the cancer cells was analyzed by immunohistochemistry and quantified using the Immunoreactive Score (IRS). Outcomes of the chemotherapy were measured by the residual cancer burden (RCB) system.
Results
CREB3L1 expression levels in TNBC responsive to doxorubicin-based chemotherapy (RCB class 0-2) were significantly higher than that in resistant cancers (RCB class 3) (unpaired two-tailed t test, p = 0.0005; Statistical power 99.8 at 95% confidence level). All cancers expressing higher levels of CREB3L1 (IRS 4-12) responded to doxorubicin-based chemotherapy, whereas all cancers resisting the treatment expressed lower levels of CREB3L1 (IRS 0-3).
Conclusions
These results suggest that CREB3L1 expression level may be used as a biomarker to identify TNBC patients who are more likely to benefit from doxorubicin-based chemotherapy.
Similar content being viewed by others
Background
Breast cancer can be divided into subgroups based on expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2, which are growth factor receptors that are crucial for proliferation of cancer cells. While targeted therapies antagonizing these receptors have significantly improved the treatment of breast cancers expressing these receptors, treating those that express none of the three receptors, namely triple negative breast cancer (TNBC), remains a daunting challenge [1]. Chemotherapy is still the mainstream treatment for these cancers. Nearly all chemotherapy regimens used to treat TNBC contain multiple drugs, and the majority of those include doxorubicin due to its effectiveness demonstrated through decades of clinical practice [2]. However, the response rate of doxorubicin-based chemotherapy is only 40–70%, primarily because there is no biomarker capable of identifying patients who will respond to doxorubicin [3,4,5]. Considering the severe cardiac toxicity caused by the side effect of doxorubicin [6], establishment of such a biomarker will not only improve the response rate of doxorubicin-based chemotherapy, but also reduce the toxicity of the treatment by avoiding application of doxorubicin to patients who are unlikely to respond to doxorubicin.
Doxorubicin has been demonstrated to induce DNA damage by inhibiting topoisomerase II, but whether this activity is associated with its ability to suppress cancer cell proliferation is still under debate [7, 8]. We have recently reported that doxorubicin blocks proliferation of cancer cells through activation of a transcription factor called cAMP response element binding protein 3-like 1 (CREB3L1) [9]. Unlike most transcription factors, CREB3L1 is synthesized as an inactive transmembrane precursor, with the transcriptionally active N-terminal domain anchored to membranes via a single transmembrane helix [10]. Doxorubicin stimulates proteolytic cleavage of CREB3L1, releasing the N-terminal domain of the protein from membranes, allowing it to enter nucleus where it activates transcription of genes that inhibit cell proliferation [9, 10]. As a result, doxorubicin inhibited proliferation of tumor cells cultured in vitro that express CREB3L1 but not those in which expression of the gene was inhibited, even though DNA damage induced by the drug was indistinguishable among these cells [9]. Using a mouse xenograft model of human renal cell carcinoma reported to maintain drug sensitivities displayed in patients [11], we previously reported that doxorubicin at a dose lower than that typically applied to human patients shrank tumors expressing high levels of CREB3L1 but not those expressing low levels of the protein [12]. Notably, at this low dose doxorubicin did not induce DNA damage in the xenograft tumors regardless of their CREB3L1 expression levels [12]. These in vitro and in vivo results suggest that doxorubicin inhibits tumor cell proliferation through proteolytic activation of CREB3L1 but not DNA damage. These findings demonstrate that CREB3L1 expression in cancer cells could be a key determinant for their sensitivity to doxorubicin.
In the current study, we performed a retrospective analysis on core biopsy samples taken from TNBC patients before chemotherapy to measure CREB3L1 expression levels in cancer cells, and to determine the relationship between CREB3L1 expression levels and outcomes of doxorubicin-based chemotherapy. The results showed that CREB3L1 expression levels in cancers that responded to chemotherapy were significantly higher than those that resisted the treatment. Our findings suggest that high levels of CREB3L1 expression in cancer cells could serve as a predictive biomarker to identify TNBC patients who are likely to respond to doxorubicin-based chemotherapy.
Methods
Materials
We obtained a rabbit polyclonal antibody against CREB3L1 from Proteintech (Cat# 11235-2-AP); peroxidase-conjugated secondary antibodies from Jackson ImmunoResearch; and rabbit anti-Actin from Sigma-Aldrich. A mouse monoclonal antibody against human CREB3L1 (10H1) was generated by immunizing mice with synthesized polypeptides corresponding to amino acids 7-41 of human CREB3L1 [12]. Formalin fixed and paraffin embedded tissue sections of TNBC core biopsies were obtained from UT Southwestern University Hospitals. Written informed consent for participation in the study was obtained from participants or, where participants were children, a parent or guardian. All animal experiments reported previously that were mentioned in the manuscript were approved by an institutional animal care and use committee of UT Southwestern Medical Center (APN 2011-0192). No animal experiment is conducted in the current study.
Verification of antibodies
Human hepatoma Huh7 cells stably transfected with a control shRNA or that targeting CREB3L1 were cultured as described previously [9]. Cells were lysed in buffer A (25 mM Tris-HCl pH = 7.2, 150 mM NaCl, and 1% NP-40) supplemented with cOmplete ULTRA protease inhibitor tablets (Roche) according to manufactures’ direction, analyzed by SDS-PAGE (10% acrylamide) followed by immunoblot analysis with the indicated antibodies (1:4000 dilution for 10H1, 1:1000 dilution for Proteintech 11235-2-AP, and 1:10,000 dilution for anti-Actin). Bound antibodies were visualized with a peroxidase-conjugated secondary antibody using the SuperSignal ECL-HRP substrate system (Pierce).
Detection of CREB3L1 expression by Immunohistochemistry (IHC)
Eighteen TNBC core biopsy slides were subjected to IHC analysis with anti-CREB3L1 (10H1) to determine CREB3L1 expression in the tumors. Briefly, paraffin-embedded sections were treated with xylenes, washed sequentially with 95, 70, 50, 30% ethanol followed by a pure water wash. Endogenous peroxidase activity was blocked by incubating the slides with 3% hydrogen peroxide for 5 minutes. After washing with PBS, antigen retrieval was performed using the Retriever 2100 with Buffer U (Electron Microscopy Sciences) according to the manufactures’ directions. Sections were then blocked with Blocking Buffer (1% Horse Serum in PBS) for 1 hour, incubated with 17 μg/ml 10H1 overnight in Blocking Buffer at 4°C, washed three times with PBS, and developed with the DAB staining procedure using the Vectastain Elite ABC Kit and DAB Peroxidase (HRP) Substrate Kit (Vector Laboratories). Nuclei counterstaining was performed by incubating the slides with hematoxylin for 5 sec. The IHC results were captured by bright field images taken by a Zeiss Observer Z1 microscope using AxioVision software, and quantified through ImmunoReactive Score (IRS) as previously described [13]. The IRS quantification was verified by Dr. Yan Peng, a board-certified pathologist.
Clinical characterization of tumors
Patient responses to doxorubicin-based chemotherapy applied after core biopsies had been taken were quantified through the residual cancer burden (RCB) system as previously described [14]. Expression of Ki67 in the tumor cells was quantified as previously reported [15].
Statistical Analysis
We retrospectively analyzed TNBC patients whose core biopsies taken before doxorubicin-based chemotherapy were available, and whose response to the chemotherapy was recorded. We analyzed 18 samples meeting the requirement. A student t-test (two tailed, two sample unequal variance) was performed on results shown in Fig. 3a. Power calculation was performed on results shown in Fig. 3a at 95% confidence interval using a one-tail test. Pearson analysis was performed on results shown in Fig. 3b.
Results
A specific antibody against CREB3L1 without significant cross reactivity is required for IHC analysis of CREB3L1. The monoclonal antibody 10H1 we raised previously [12] is suitable for this application: On immunoblot the antibody detected only the precursor and cleaved form of CREB3L1 (Fig. 1, lane 1) in Huh7 cells stably transfected with a control shRNA that expressed CREB3L1, but detected no protein in Huh7 cells stably transfected with a shRNA targeting CREB3L1 that inhibited expression of the mRNA by more than 90% [9] (Fig. 1, lane 2). In contrast to 10H1, a commercially available anti-CREB3L1 stated suitable for IHC had much more cross reactivity (Fig. 1, lanes 3 and 4).
10H1 is a highly specific antibody against CREB3L1. On day 0, Huh7 cells stably transfected with a control shRNA or that targeting CREB3L1 were seeded at 4×105 cells per 60 mm dish. On day 1, cells were harvest, and cell lysates were subjected to immunoblot analysis with antibodies reacting against CREB3L1 (10H1 or Proteintech Cat# 11235-2-AP) and Actin
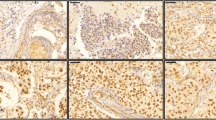
We then obtained unstained slides of core biopsies taken from TNBC patients before they were subjected to doxorubicin-based chemotherapy, and used 10H1 to measure CREB3L1 expression in the tumor cells through IHC. Some typical results of the IHC analysis were shown in Fig. 2, which include TNBC with no detectable expression (Fig. 2a), with heterogeneous expression (Fig. 2b), or with homogenous high levels of expression of CREB3L1 (Fig. 2c).
We quantified CREB3L1 expression in cancer cells of all patients before doxorubicin-based chemotherapy through the IRS system, and measured their response to the following chemotherapy through the RCB system. In the RCB system, the higher the score, the more resistant the tumor to chemotherapy [14]. Based on the RCB scores, the tumors are divided into 4 classes, with class 0, 1-2, and 3 refers to completely responsive, partially responsive, and completely resistant tumors, respectively [14]. The results were summarized in Table 1. According to our hypothesis, higher levels of CREB3L1 expression should render cancer cells more sensitive to doxorubicin. Consistent with this hypothesis, we observed that the average IRS value of tumors that completely or partially responded to the chemotherapy (RCB classes 0-2) was 8 times higher than that of the tumors resistant to the treatment (RCB class 3) (4.8 vs 0.6) (Fig. 3a). This difference is statistically significant indicated by the student t-test (p = 0.0005) and power analysis (statistical power 99.8% assuming a 95% confidence interval). Likewise, the degree of resistance to the chemotherapy measured by increased RCB scores was negatively correlated with CREB3L1 expression levels (Pearson analysis, R = -0.54, p = 0.02) (Fig. 3b).
Higher levels of CREB3L1 expression in TNBC demonstrate an improved response to doxorubicin-based chemotherapy. a A student t-test (two tailed, two sample unequal variance) was performed to compare CREB3L1 IRS values in tumors responded to doxorubicin-based chemotherapy (RCB Class 0-2, responder) versus those resisted the treatment (RCB Class 3, non-responder). b Pearson analysis was performed to determine the correlation between CREB3L1 IRS values and RCB scores of the tumors. The hollow dot indicates two samples at the same location on the plot. c Percentage of cases with indicated RCB class in tumors with lower expression of CREB3L1 (IRS value 0-3) and those with higher expression of the protein (IRS value 4-12)
To determine whether CREB3L1 expression levels can serve as a biomarker to predict outcomes of doxorubicin-based chemotherapy, we separated the tumors into two groups: One with lower (IRS ranges from 0 to 3) and the other with higher (IRS ranges from 4 to 12) levels of CREB3L1 expression. We observed that all tumors resistant to doxorubicin-based chemotherapy fell in the group with lower expression of CREB3L1 (Fig. 3c). As a result, only 50% of the tumors in this group responded to the chemotherapy (Fig. 3c). In contrast, all tumors with higher expression of CREB3L1 were responsive to the chemotherapy (Fig. 3c).
Discussion
We have previously reported that doxorubicin inhibits proliferation of tumor cells through proteolytic activation of CREB3L1, and CREB3L1 expression determines the sensitivity of tumors to doxorubicin in various in vitro and in vivo experimental systems [9, 12]. The current study demonstrates the clinical importance of the finding by establishing CREB3L1 expression as a potential biomarker capable of identifying TNBC patients who are likely to benefit from doxorubicin-based chemotherapy. We show that tumors with higher levels of CREB3L1 expression (IRS value ≥ 4) all responded to doxorubicin-based chemotherapy. Thus, applying such chemotherapy only to patients with higher levels of CREB3L1 expression should markedly increase the response rate of the treatment. We also observe that all tumors resistant to doxorubicin-based chemotherapy had lower levels of CREB3L1 expression (IRS value ≤ 3). However, some tumors with lower levels of CREB3L1 expression did respond to the chemotherapy. The most likely explanation for this observation is that TNBC patients are not treated with doxorubicin alone, and these tumors may be sensitive to other drugs included in the chemotherapy regimens such as Cytoxan and Taxol. If this is the case, then for tumors with lower levels of CREB3L1 expression, replacing doxorubicin with other chemotherapeutic reagents could prevent unnecessary cardiac toxicity associated with doxorubicin [6] without affecting the response rate of the treatment.
The current study, in which only 18 samples were analyzed, was originally designed as a pilot study to estimate the sample size required to reach statistical significance. Surprisingly, our small sample number was sufficient for us to draw the conclusion that CREB3L1 expression levels in tumors responsive to doxorubicin-based chemotherapy were higher than those resistant to the treatment. This is because the difference in CREB3L1 expression levels between the two groups of the tumors was so large (~ 8 fold) that even with such a limited sample size, statistical significance was achieved (p = 0.0005) though a simple student t-test. More importantly, power analysis, which has been established as the standard for sample size estimation in clinical studies [16], demonstrated that our statistic power (99.8%) was much higher than the value (80%) considered statistically significant in most clinical studies. Despite the small sample size, our sample selection was not skewed in terms of response of the tumors to chemotherapy, as the percentage of cases completely responsive or resistant to the therapy in our samples was similar to that in clinical trials involving hundreds of patients [3, 17]. Thus, adding more samples to this retrospect study will not further improve statistical significance in a clinically meaningful manner. Instead, this study should serve as a base for future prospective studies to firmly establish CREB3L1 as a clinical biomarker to identify TNBC patients who are most likely to respond to doxorubicin-based chemotherapy.
The current study reveals that CREB3L1 expression varies widely among different TNBC tumors. About 50% of the tumors we examined had low levels of CREB3L1 expression (IRS value ≤ 3). Previous reports have indicated that expression of CREB3L1 is frequently epigenetically silenced through DNA methylation in various cancers including breast cancer [18,19,20,21]. We have previously reported that azacitidine, a drug that inhibits DNA methylation [22], relieved epigenetic silencing of CREB3L1 [18]. In cultured MCF-7 cells in which CREB3L1 expression is barely detectable [9], azacitidine markedly increase the sensitivity of the cells to doxorubicin [23]. It will be interesting to determine whether addition of azacitidine to doxorubicin-based chemotherapy regimens may improve the treatment outcomes for TNBC with lower levels of CREB3L1 expression.
Conclusion
The current study demonstrates that TNBC tumors with higher expression of CREB3L1 have a higher response rate to doxorubicin-based chemotherapy compared to those with lower expression of the protein. These results suggest that CREB3L1 expression level may serve as a biomarker to identify TNBC patients who are more likely to benefit from doxorubicin-based chemotherapy.
Abbreviations
- CREB3L1:
-
Cyclic adenosine monophosphate response element binding protein 3 like-1
- IHC:
-
Immunohistochemistry
- IRS:
-
ImmunoReactive score
- RCB:
-
Residual cancer burden
- TNBC:
-
Triple negative breast cancer
References
Peng Y, Butt YM, Chen B, Zhang X, Tang P. Update on immunohistochemical analysis in breast lesions. Arch Pathol Lab Med. 2017;141(8):1033–51.
Rampurwala MM, Rocque GB, Burkard ME. Update on adjuvant chemotherapy for early breast cancer. Breast Cancer. 2014;8:125–33.
Nabholtz J-MA, Reese DM, Lindsay M-A, Riva A. Docetaxel in the treatment of breast cancer: An update on recent studies. Semin Oncol. 2002;29(3):28–34.
Fisher B, Jeong J-H, Anderson S, Wolmark N. Treatment of axillary lymph node–negative, estrogen receptor–negative breast cancer: Updated findings from national surgical adjuvant breast and bowel project clinical trials. J Natl Cancer Inst. 2004;96(24):1823–31.
Kaufman PA. Paclitaxel and anthracycline combination chemotherapy for metastatic breast cancer. Semin Oncol. 1999;26:39–46.
Chatterjee K, Zhang J, Honbo N, Karliner JS. Doxorubicin Cardiomyopathy. Cardiology. 2010;115(2):155–62.
Patel AG, Kaufmann SH. How does doxorubicin work? eLife. 2012;1
Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol. 1999;57:727–41.
Denard B, Lee C, Ye J. Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1. eLife Sci. 2012;1 https://doi.org/10.7554/eLife.00090.
Denard B, Seemann J, Chen Q, Gay A, Huang H, Chen Y, Ye J. The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host Microbe. 2011;10:65–74.
Sivanand S, Peña-Llopis S, Zhao H, Kucejova B, Spence P, Pavia-Jimenez A, Yamasaki T, McBride DJ, Gillen J, Wolff NC, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4(137):137ra175.
Denard B, Pavia-Jimenez A, Chen W, Williams NS, Naina H, Collins R, Brugarolas J, Ye J. Identification of CREB3L1 as a biomarker predicting doxorubicin treatment outcome. PLoS ONE. 2015;10(6):e0129233.
Fedchenko N, Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue – a review. Diagn Pathol. 2014;9:221.
Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, Assad L, Poniecka A, Hennessy B, Green M, et al. Measurement of Residual Breast Cancer Burden to predict survival after neoadjuvant chemotherapy. J clin Oncol. 2007;25(28):4414–22.
Han JS, Cao D, Molberg KH, Sarode VR, Rao R, Sutton LM, Peng Y. Hormone receptor status rather than HER2 status is significantly associated with increased Ki-67 and p53 expression in triple-negative breast carcinomas, and high expression of Ki-67 but not p53 Is significantly associated with axillary nodal metastasis in triple-negative and high-grade non–triple-negative breast carcinomas. Am J Clin Pathol. 2011;135(2):230–7.
Whitley E, Ball J. Statistics review 4: Sample size calculations. Crit Care. 2002;6(4):335–41.
Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–81.
Chen Q, Denard B, Huang H, Ye J. Epigenetic silencing of antiviral genes renders clones of Huh-7 cells permissive for Hepatitis C virus replication. J Virol. 2013;87(1):659–65.
Rose M, Schubert C, Dierichs L, Gaisa NT, Heer M, Heidenreich A, Knüchel R, Dahl E. OASIS/CREB3L1 is epigenetically silenced in human bladder cancer facilitating tumor cell spreading and migration in vitro. Epigenetics. 2014;9(12):1626–40.
Steenbergen RDM, Ongenaert M, Snellenberg S, Trooskens G, van der Meide WF, Pandey D, Bloushtain-Qimron N, Polyak K, Meijer CJLM, Snijders PJF, et al. Methylation-specific digital karyotyping of HPV16E6E7-expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J Pathol. 2013;231(1):53–62.
Ward AK, Mellor P, Smith SE, Kendall S, Just NA, Vizeacoumar FS, Sarker S, Phillips Z, Alvi R, Saxena A, et al. Epigenetic silencing of CREB3L1 by DNA methylation is associated with high-grade metastatic breast cancers with poor prognosis and is prevalent in triple negative breast cancers. Breast Cancer Res. 2016;18:12.
Sato T, Issa J-PJ, Kropf P. DNA hypomethylating drugs in cancer therapy. Cold Spring Harb Perspect Med. 2017;7(5):a026948. https://doi.org/10.1101/cshperspect.a026948.
Khan GN, Kim EJ, Shin TS, Lee SH. Azacytidine-induced chemosensitivity to doxorubicin in human breast cancer MCF7 Cells. Anticancer Res. 2017;37(5):2355–64.
Acknowledgement
We thank Drs. Michael S. Brown and Joseph L. Goldstein for their support and advice; Lisa Beatty, Lauren Valsin and Ijeoma Onwuneme for assistance with tissue culture; and JungYeon Kim for technical assistance.
Funding
This study was supported by grants from the National Institutes of Health (GM-116106 and HL-20948) and the Welch Foundation (I-1832). These grants provided financial supports for the study but did not impose restrictions on design of the study and collection, analysis, interpretation of data, or writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
BD designed and performed experiments, analyzed data, and wrote the manuscript. SJ analyzed data and wrote the manuscript. YP designed experiments, gathered samples, wrote the manuscript, and analyzed data. JY designed experiments, analyzed data and wrote the manuscript. All authors have read and approved the manuscript
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Informed consent to participate in the study was obtained from participants. The use of the patient materials and information has been approved by the University of Texas Southwestern Medical Center Institutional Review Board (IRB# STU032011-117).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Denard, B., Jiang, S., Peng, Y. et al. CREB3L1 as a potential biomarker predicting response of triple negative breast cancer to doxorubicin-based chemotherapy. BMC Cancer 18, 813 (2018). https://doi.org/10.1186/s12885-018-4724-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-018-4724-8