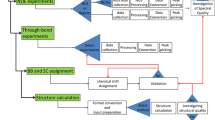
Abstract
The heterogeneous array of software tools used in the process of protein NMR structure determination presents organizational challenges in the structure determination and validation processes, and creates a learning curve that limits the broader use of protein NMR in biology. These challenges, including accurate use of data in different data formats required by software carrying out similar tasks, continue to confound the efforts of novices and experts alike. These important issues need to be addressed robustly in order to standardize protein NMR structure determination and validation. PDBStat is a C/C++ computer program originally developed as a universal coordinate and protein NMR restraint converter. Its primary function is to provide a user-friendly tool for interconverting between protein coordinate and protein NMR restraint data formats. It also provides an integrated set of computational methods for protein NMR restraint analysis and structure quality assessment, relabeling of prochiral atoms with correct IUPAC names, as well as multiple methods for analysis of the consistency of atomic positions indicated by their convergence across a protein NMR ensemble. In this paper we provide a detailed description of the PDBStat software, and highlight some of its valuable computational capabilities. As an example, we demonstrate the use of the PDBStat restraint converter for restrained CS-Rosetta structure generation calculations, and compare the resulting protein NMR structure models with those generated from the same NMR restraint data using more traditional structure determination methods. These results demonstrate the value of a universal restraint converter in allowing the use of multiple structure generation methods with the same restraint data for consensus analysis of protein NMR structures and the underlying restraint data.






Similar content being viewed by others
Abbreviations
- ACO:
-
Dihedral angle constraint
- CNSw:
-
Protocol using the crystallography and NMR software (CNS) package for restrained structure refinement in explicit water solvent
- DAOP:
-
Dihedral angle order parameter
- CS:
-
Chemical shift
- rCS-Rosetta:
-
Restrained chemical shift-directed Rosetta
- RDC:
-
Residual dipolar coupling
- SVD:
-
Singular value decomposition
- RMSD:
-
Root mean squared deviation
References
Bahrami A, Assadi AH, Markley JL, Eghbalnia HR (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol 5:e1000307
Baran MC, Huang YJ, Moseley HN, Montelione GT (2004) Automated analysis of protein NMR assignments and structures. Chem Rev 104:3541–3556
Baran MC, Moseley HN, Aramini JM, Bayro MJ, Monleon D, Locke JY, Montelione GT (2006) SPINS: a laboratory information management system for organizing and archiving intermediate and final results from NMR protein structure determinations. Proteins 62:843–851
Bassolino-Klimas D, Tejero R, Krystek SR, Metzler WJ, Montelione GT, Bruccoleri RE (1996) Simulated annealing with restrained molecular dynamics using a flexible restraint potential: theory and evaluation with simulated NMR constraints. Protein Sci 5:593–603
Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66:778–795
Bhattacharya A, Wunderlich Z, Monleon D, Tejero R, Montelione GT (2008) Assessing model accuracy using the homology modeling automatically (HOMA) software. Proteins 70:105–118
Braun W, Go N (1985) Calculation of protein conformations by proton-proton distance constraints: a new efficient algorithm. J Mol Biol 186:611–626
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr D 54:905–921
Cornilescu G, Marquardt JL, Ottiger M, Bax A (1998) Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 120:6836–6837
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Doreleijers JF, Raves ML, Rullmann T, Kaptein R (1999) Completeness of NOEs in protein structure: a statistical analysis of NMR data. J Biomol NMR 14:123–132
Doreleijers JF, Mading S, Maziuk D, Sojourner K, Yin L, Zhu J, Markley JL, Ulrich EL (2003) BioMagResBank database with sets of experimental NMR constraints corresponding to the structures of over 1400 biomolecules deposited in the Protein Data Bank. J Biomol NMR 26:139–146
Doreleijers JF, Sousa da Silva AW, Krieger E, Nabuurs SB, Spronk CA, Stevens TJ, Vranken WF, Vriend G, Vuister GW (2012a) CING: an integrated residue-based structure validation program suite. J Biomol NMR 54:267–283
Doreleijers JF, Vranken WF, Schulte C, Markley JL, Ulrich EL, Vriend G, Vuister GW (2012b) NRG-CING: integrated validation reports of remediated experimental biomolecular NMR data and coordinates in wwPDB. Nucleic Acids Res 40:D519–D524
Güntert P, Braun W, Wüthrich K (1991) Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol 217:517–530
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298
Han B, Liu Y, Ginzinger SW, Wishart DS (2011) SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR 50:43–57
Havel TF, Wüthrich K (1985) An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol 182:281–294
Hendrickx PM, Gutmanas A, Kleywegt GJ (2013) Vivaldi: visualizaton and validation of biomacromolecular NMR structures from the PDB. Proteins 81:583–591
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227
Huang YJ, Moseley HN, Baran MC, Arrowsmith C, Powers R, Tejero R, Szyperski T, Montelione GT (2005a) An integrated platform for automated analysis of protein NMR structures. Methods Enzymol 394:111–141
Huang YJ, Powers R, Montelione GT (2005b) Protein NMR recall, precision and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc 127:1665–1674
Huang YJ, Tejero R, Powers R, Montelione GT (2006) A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 62:587–603
Huang YJ, Rosato A, Singh G, Montelione GT (2012) RPF: a quality assessment tool for protein NMR structures. Nucleic Acids Res 40:W542–W546
Hyberts SG, Goldberg MS, Havel TF, Wagner G (1992) The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci 1:736–751
Kabsch W (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr A 32:922–923
Kabsch W (1978) A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr A 34:827–828
Kirchner DK, Güntert P (2011) Objective identification of residue ranges for the superposition of protein structures. BMC Bioinformatics 12:170
Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D (2012) Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci USA 109:10873–10878
Liu G, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton TA, Arrowsmith CH, Montelione GT, Szyperski T (2005) NMR data collection and analysis protocol for high-throughput protein structure determination. Proc Natl Acad Sci USA 102:10487–10492
Losonczi JA, Andrec M, Fischer MW, Prestegard JH (1999) Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson 138:334–342
Mao B, Guan R, Montelione GT (2011) Improved technologies now routinely provide protein NMR structures useful for molecular replacement. Structure 19:757–766
Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes B, Wright P, Wüthrich K (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl Chem 70:117–142
Moseley HN, Montelione GT (1999) Automated analysis of NMR assignments and structures for proteins. Curr Opin Struct Biol 9:635–642
Moseley HN, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol 339:91–108
Moseley HN, Sahota G, Montelione GT (2004) Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR 28:341–355
Nabuurs SB, Spronk CA, Vuister GW, Vriend G (2006) Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. PLoS Comput Biol 2:e9
Nilges M (1995) Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J Mol Biol 245:645–660
Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D (2010) NMR structure determination for larger proteins using backbone-only data. Science 327:1014–1018
Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA (2009) Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins 75:147–167
Rohl CA, Strauss CE, Misura KM, Baker D (2004) Protein structure prediction using Rosetta. Methods Enzymol 383:66–93
Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Güntert P, He Y, Herrmann T, Huang YJ, Jaravine V, Jonker HR, Kennedy MA, Lange OF, Liu G, Malliavin TE, Mani R, Mao B, Montelione GT, Nilges M, Rossi P, van der Schot G, Schwalbe H, Szyperski TA, Vendruscolo M, Vernon R, Vranken WF, de Vries S, Vuister GW, Wu B, Yang Y, Bonvin AM (2012) Blind testing of routine, fully automated determination of protein structures from NMR data. Structure 20:227–236
Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160:65–73
Snyder DA, Montelione GT (2005) Clustering algorithms for identifying core atom sets and for assessing the precision of protein structure ensembles. Proteins 59:673–686
Struyf A, Hubert M, Rousseeuw P (1996) Clustering in an object-oriented environment. J Stat Softw 1:1–30
Tejero R, Bassolino-Klimas D, Bruccoleri RE, Montelione GT (1996) Simulated annealing with restrained molecular dynamics using CONGEN: energy refinement of the NMR solution structures of epidermal and type-α transforming growth factors. Protein Sci 5:578–592
Valafar H, Prestegard JH (2004) REDCAT: a residual dipolar coupling analysis tool. J Magn Reson 167:228–241
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Williams T, Kelley C (2011) Gnuplot 4.5: an interactive plotting program. http://gnuplot.info
Williamson MP, Havel TF, Wüthrich K (1985) Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol 182:295–315
Zemla A (2003) LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res 31:3370–3374
Zimmerman DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol 269:592–610
Zweckstetter M, Bax A (2000) Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J Am Chem Soc 122:3791–3792
Acknowledgments
We thank all the members of the NMR groups of the Northeast Structural Genomics Consortium who contributed constructive criticisms and test data sets used in the development of PDBStat. Special thanks to C. Arrowsmith, J. Cort, A. Eletsky, L. Fella, Y. J. Huang, A. Lemak, M. Kennedy, G. Liu, J. Prestegard, T. Ramelot, A. Rosato, G.V.T. Swapna, T. Szyperski, Y. Tang, and B. Wu for useful discussions. This work was supported by a Grant from the Protein Structure Initiative of the National Institutes of Health (U54-GM094597). RT also acknowledges suppport from CONSOLIDER INGENIO CSD2010-00065 and Generalitat Valenciana PROMETEO 2011/008. DS also acknowledges support from the Research Corporation for Science Advancement, College Cottrell Grant, Award #19803.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tejero, R., Snyder, D., Mao, B. et al. PDBStat: a universal restraint converter and restraint analysis software package for protein NMR. J Biomol NMR 56, 337–351 (2013). https://doi.org/10.1007/s10858-013-9753-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9753-7