Abstract
The J-UNIO (JCSG protocol using the software UNIO) procedure for automated protein structure determination by NMR in solution is introduced. In the present implementation, J-UNIO makes use of APSY-NMR spectroscopy, 3D heteronuclear-resolved [1H,1H]-NOESY experiments, and the software UNIO. Applications with proteins from the JCSG target list with sizes up to 150 residues showed that the procedure is highly robust and efficient. In all instances the correct polypeptide fold was obtained in the first round of automated data analysis and structure calculation. After interactive validation of the data obtained from the automated routine, the quality of the final structures was comparable to results from interactive structure determination. Special advantages are that the NMR data have been recorded with 6–10 days of instrument time per protein, that there is only a single step of chemical shift adjustments to relate the backbone signals in the APSY-NMR spectra with the corresponding backbone signals in the NOESY spectra, and that the NOE-based amino acid side chain chemical shift assignments are automatically focused on those residues that are heavily weighted in the structure calculation. The individual working steps of J-UNIO are illustrated with the structure determination of the protein YP_926445.1 from Shewanella amazonensis, and the results obtained with 17 JCSG targets are critically evaluated.




Similar content being viewed by others
References
Atreya HS, Sahu SC, Chary KVR, Govil G (2000) A tracked approach for automated NMR assignments in proteins (TATAPRO). J Biomol NMR 17:125–136
Bartels C, Güntert P, Billeter M, Wüthrich K (1997) GARANT—a general algorithm for resonance assignment in multidimensional nuclear magnetic resonance spectra. J Comput Chem 18:139–149
Cavanagh J, Fairbrother WJ, Rance M, Palmer AG III, Skelton NJ (2007) Protein NMR spectroscopy: principles and practice, 2nd edn. Elsevier Academic Press, Amsterdam
Crippen GM, Rousaki A, Revington M, Zhang Y, Zuiderweg ERP (2010) SAGA: rapid automatic mainchain NMR assignment for large proteins. J Biomol NMR 46:281–298
DeMarco A, Wüthrich K (1976) Digital filtering with a sinusoidal window function: an alternative technique for resolution enhancement in FT NMR. J Magn Reson 24:201–204
Elsliger MA, Deacon A, Godzik A, Lesley S, Wooley J, Wüthrich K, Wilson IA (2010) The JCSG high-throughput structural biology pipeline. Acta Cryst F 66:1137–1142
Fiorito F, Herrmann T, Damberger FF, Wüthrich K (2008) Automated amino acid side-chain NMR assignment of proteins using 13C- and 15N-resolved [1H,1H]-spectra. J Biomol NMR 42:23–33
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298
Herrmann T, Güntert P, Wüthrich K (2002a) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227
Herrmann T, Güntert P, Wüthrich K (2002b) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24:171–189
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102(31):10876–10881
Hiller S, Wider G, Wüthrich K (2008) APSY-NMR with proteins: practical aspects and backbone assignment. J Biomol NMR 42:179–195
Ikeya T, Jee J-G, Shigemitsu Y, Hamatsu J, Mishima M, Ito Y, Kainosho M, Güntert P (2011) Exclusively NOESY-based automated NMR assignment and structure determination of proteins. J Biomol NMR 50:137–146
Jaudzems K, Geralt M, Serrano P, Mohanty B, Horst R, Pedrini B, Elsliger MA, Wilson IA, Wüthrich K (2010) NMR structure of the protein NP_247299.1: comparison with the crystal structure. Acta Cryst F 66:1367–1380
Keller R (2004) CARA: computer aided resonance assignment. http://cara.nmr.ch/
Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55
Kraulis PJ (1994) Protein three-dimensional structure determination and sequence-specific assignment of 13C and 15N-separated NOE data. J Mol Biol 243:696–728
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Lemak A, Steren CA, Arrowsmith CH, Llinas M (2008) Sequence specific resonance assignment via multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR 41:29–41
Lescop E, Brutscher B (2009) Highly automated protein backbone resonance assignment within a few hours: the «BATCH» strategy and software package. J Biomol NMR 44:43–57
Lesley S, Kuhn P, Godzik A, Deacon A, Mathews I, Kreusch A, Spraggon G, Klock H, McMullan D, Shin T, Vincent J, Robb A, Brinen L, Miller M, McPhillips T, Miller M, Scheibe D, Canaves J, Guda C, Jaroszewski L, Selby T, Elsliger MA, Wooley J, Taylor S, Hodgson K, Wilson IA, Schultz P, Stevens R (2002) Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci USA 99:11664–11669
Lüthy R, Bowie J, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85
Metzler W, Constantine K, Friedrichs M, Bell A, Ernst E, Lavoie T, Mueller L (1993) Charecterization of the three-dimensional solution structure of human profilin: 1H, 13C, and 15N NMR assignments and global folding pattern. Biochemistry 32:13818–13829
Mohanty B, Serrano P, Pedrini B, Jaudzems K, Geralt M, Horst R, Herrmann T, Elsliger ME, Wilson IA, Wüthrich K (2010) NMR structure of the protein NP_247299.1: comparison with the crystal structure. Acta Cryst F 66:1381–1392
Moseley HN, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Meth Enzym 399:91–108
Page R, Peti W, Wilson IA, Stevens RC, Wüthrich K (2005) NMR screening and crystal quality of bacterially expressed prokaryotic and eukaryotic proteins in a structural genomics pipeline. Proc Natl Acad Sci USA 102(6):1901–1905
Peti W, Page R, Moy K, O’Neil-Johnson M, Wilson IA, Stevens RC, Wüthrich K (2005) Towards miniaturization of a structural genomics pipeline using macro-expression and microcoil NMR. J Struct Funct Genomics 6:259–267
Schmucki R, Yokohama S, Güntert P (2008) Automated assignment of NMR chemical shifts using peak-particle dynamics simulation with the DYNASSIGN algorithm. J Biomol NMR 43:97–109
Serrano P, Pedrini B, Geralt M, Jaudzems K, Mohanty B, Horst R, Herrmann T, Elsliger MA, Wilson IA, Wüthrich K (2010) Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites. Acta Cryst F 66(10):1392–1405
Staykova DK, Fredriksson J, Bermel W, Billeter M (2008) Assignment of protein NMR spectra based on projections, multi-way decomposition and a fast correlation approach. J Biomol NMR 42:87–97
Volk J, Herrmann T, Wüthrich K (2008) Automated sequence-specific protein NMR assignment using the memetic algorithm MATCH. J Biomol NMR 41:127–138
Wishart D, Sykes B (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:135–140
Wüthrich K (1986) NMR of proteins and nucleic acids. Wiley, New York
Wüthrich K (2010) NMR in a crystallography-based high-throughput protein structure-determination environment. Acta Cryst F 66:1365–1366
Zimmermann DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol 269:592–610
Acknowledgments
The following financial support is acknowledged: Swiss National Science Foundation and ETH Zürich through the NCCR Structural Biology; Swiss National Science Foundation for a Fellowship to BP (PA00A–104097/1); NIH, National Institute of General Medical Services, Protein Structure Initiative, Grants U54 GM094586 and U54 GM074898 (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Science or the National Institutes of Health). KW is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Pedro Serrano and Bill Pedrini contributed equally to this work.
Appendix: Validation of J-UNIO NMR structures
Appendix: Validation of J-UNIO NMR structures
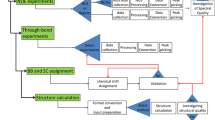
Our validation strategy makes use of quantitative criteria to qualify the Structure V (Fig. 1), including the publically available tools Procheck (Laskowski et al. 1993), Verify3D (Lüthy et al. 1992) and the PDB validation suite. In-house threshold values for acceptance of the individual criteria (Table 1) were established based on past high-quality interactive protein structure determinations in our laboratory. Furthermore, some qualitative tools are used for initial checks of the final Structure V, in order to guide the spectroscopist during the early stages of the validation procedure, and additional tools are used to monitor the course of the automated structure determination. In the following we comment on the validation tools represented in Table 1, and then on the additional criteria.
A first criterion considered in Table 1 enables an evaluation of the input for the protein structure calculation, i.e., we request that the number of long-range NOE constraints per residue must be higher than the threshold of five. In our experience, satisfying this sole criterion is sufficient to document that nearly complete chemical shift assignments have been obtained and that there is also a dense network of sequential and medium-range NOE distance constraints, thus qualifying an input for the structure calculation that is of high overall quality.
A second group of criteria is used to document acceptable convergence of the structure calculation, with small residual violations of the experimental input data and small distortions of the covalent structure geometry. These are the residual target function value, the number of residual NOE distance constraint violations, the number of residual dihedral angle violations, and the RMSD from standard covalent structure geometry.
In a third group of criteria, the precision of the Structure V (Fig. 1) is characterized by RMSDs to the mean coordinates of the bundle of conformers (Fig. 4b) calculated for the backbone heavy atoms and all heavy atoms, respectively. In addition, we introduce the “core precision” as the all-heavy-atom RMSD calculated for all the residues with solvent accessibility below 15 %. Initial experience with this parameter indicates that it is useful for comparison of the core packing in different protein structure types. The overall quality of the Structure V is monitored also by the PROCHECK global quality score, the Verify3D raw score, and the side chain planarity Z-score, with the acceptance threshold values listed in Table 1. In addition, a structure is accepted only if all criteria of the PDB validation suite are satisfied.
Additional qualitative criteria for structure validation are used to directly assess the agreement between selected raw experimental NMR data and corresponding data derived from the Structure V bundle of conformers (Fig. 4b). First, comparison of the structure-derived and the observed ring current shifts provides qualitative checks on possible local errors in amino acid side chain arrangements. The Fig. 5 shows a plot of the observed methyl hydrogen ring current shifts (RCSobs) versus the corresponding ring current shifts calculated from the atomic coordinates of the NMR structure (RCSpre) for the protein YP_926445.1. Prior to structure validation with the tools listed in Table 1, methyl groups with entries located far from the diagonal in this presentation would be singled out for further interactive analysis until a satisfactory fit is attained, or a rationale is found to explain the apparent discrepancy. Second, comparison of the regular secondary structures in Structure V and those predicted from the 13Cα and 13Cβ chemical shift values (Fig. 6) afford a check of the agreement between experimental NMR data for the polypeptide backbone and the final Structure V (Wishart and Sykes 1994), and the same applies to analysis of the agreement between experimental patterns of sequential and medium-range 1H–1H-NOEs and the locations of regular secondary structures in Structure V (Fig. 7) (Wüthrich 1986). Similar to the aforementioned handling of the ring current shift data, apparent discrepancies between the locations of regular secondary structures, the corresponding 13Cα and 13Cβ chemical shift values and/or the NOE patterns are followed up prior to the structure validation reported in Table 1.
Plot of observed methyl hydrogen ring current shifts (RCSobs) for the protein YP_926445.1 versus the corresponding ring current shifts calculated from the atomic coordinates of the NMR structure (RCSpre). RCSobs is the difference between corresponding observed and random coil chemical shifts. RCSpre was computed with the Johnson–Bovey model implemented in the software MOLMOL (Koradi et al. 1996), and the average over the 20 NMR conformers of Structure V (Fig. 4b, c) is given
Sum of the secondary 13Cα and 13Cβ chemical shifts, Δδi, in the protein YP_926445.1 plotted versus the amino acid sequence. The Δδi value for residue i represents the average over the three consecutive residues i − 1, i and i + 1: Δδi = 1/3 (ΔδC αi−1 + ΔδC αi + ΔδC αi+1 + ΔδC βi−1 + ΔδC βi + ΔδC βi+1 ) (Metzler et al. 1993). The ΔδCα and ΔδCβ values were determined with the program package UNIO-ATNOS/CANDID (Herrmann et al. 2002a, b) by subtracting the random coil shifts from the experimentally determined chemical shifts. Positive Δδi values indicate that the residue i is located in a helical structure, while a negative value indicates a location in a β-strand. The positions of the regular secondary structures in the Structure V are indicated at the top of the figure
Sequential and medium-range 1H–1H NOE constraints observed for YP_926445.1. The amino acid sequence and the regular secondary structures identified by MOLMOL (Koradi et al. 1996) in Structure V are indicated at the top. Residues are included in the regular secondary structures if the criteria are satisfied for at least 15 conformers in the bundle of 20 conformers (Fig. 4b). In the notations for the 1H–1H NOEs on the left, N, α and β indicate the HN, Hα and Hβ atoms, respectively. Sequential NOEs are indicated by continuous horizontal lines extending over the connected polypeptide segments, where thick and thin lines represent strong and weak NOEs, respectively. Medium-range NOEs are indicated by horizontal lines linking the two residues that are connected by the NOE (Wüthrich 1986)
The Table 3 lists the three principal criteria that we use to monitor the course of the calculation of Structure V with the software UNIO-ATNOS/CANDID and the simulated annealing routine of CYANA (for the initial round of calculations which result in Structure A, we only evaluate the final result obtained after cycle 7 (Herrmann et al. 2002a), since the criteria of Table 3 would be dominantly affected by the obvious limitations of the input used, as is described in the main text). The CYANA target function value must be below the threshold of 300 Å2 after the first cycle, should then monotonously adopt smaller values after cycles 2–6, and be below the threshold of 10 Å2 after cycle 7. The percentage of covalent NOEs assigned (Herrmann et al. 2002b) is automatically recorded by the ATNOS module in UNIO-ATNOS/CANDID. Obtaining high completeness of these “covalent assignments” assures robustness of the 1H–1H-NOE-based approach used by J-UNIO. Finally, checking the extent to which the NOE cross peaks in the three NOESY data sets (Fig. 1) have been assigned serves primarily to evaluate the success of the effort made for the interactive completion of the assignments from the automated routines. Rationales for choosing the rather permissible threshold of <20 % are given in the main text.
Rights and permissions
About this article
Cite this article
Serrano, P., Pedrini, B., Mohanty, B. et al. The J-UNIO protocol for automated protein structure determination by NMR in solution. J Biomol NMR 53, 341–354 (2012). https://doi.org/10.1007/s10858-012-9645-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9645-2