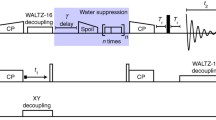
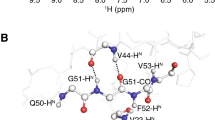
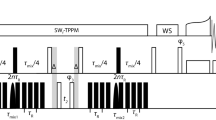
Abstract
For micro-crystalline proteins, solid-state nuclear magnetic resonance spectroscopy of perdeuterated samples can provide spectra of unprecedented quality. Apart from allowing to detect sparsely introduced protons and thereby increasing the effective resolution for a series of sophisticated techniques, deuteration can provide extraordinary coherence lifetimes—obtainable for all involved nuclei virtually without decoupling and enabling the use of scalar magnetization transfers. Unfortunately, for fibrillar or membrane-embedded proteins, significantly shorter transverse relaxation times have been encountered as compared to micro-crystalline proteins despite an identical sample preparation, calling for alternative strategies for resonance assignment. In this work we propose an approach towards sequential assignment of perdeuterated proteins based on long-range 1H/13C Cross Polarization transfers. This strategy gives rise to H/N-separated correlations involving Cα, Cβ, and CO chemical shifts of both, intra- and interresidual contacts, and thus connecting adjacent residues independent of transverse relaxation times.






Similar content being viewed by others
References
Agarwal V, Diehl A, Skrynnikov N (2006) High resolution 1H detected 1H, 13C correlation spectra in MAS solid-state NMR using deuterated proteins with selective 1H, 2H isotopic labeling of methyl groups. J Am Chem Soc 128:12620–12621
Agarwal V, Linser R, Fink U, Faelber K, Reif B (2010) Identification of hydroxyl protons, determination of their exchange dynamics, and characterization of hydrogen bonding by MAS solid-state NMR spectroscopy in a microcrystalline protein. J Am Chem Soc 132:3187–3195
Akbey Ü et al (2009a) Optimum levels of exchangeable protons in perdeuterated proteins for proton detection in MAS solid-state NMR spectroscopy. J Biomol NMR 46:67–73
Akbey Ü, Oschkinat H, van Rossum B (2009b) Double-nucleus enhanced recoupling for efficient 13C MAS NMR correlation spectroscopy of perdeuterated proteins. J Am Chem Soc 131:17054–17055
Akbey Ü, Camponeschi F, van Rossum B-J, Oschkinat H (2011) Triple resonance cross-polarization for more sensitive 13C MAS NMR spectroscopy of deuterated proteins. ChemPhysChem doi:10.1002/cphc.201100084:
Asami S, Schmieder P, Reif B (2010) High resolution 1H-detected solid-state NMR spectroscopy of protein aliphatic resonances: access to tertiary structure information. J Am Chem Soc 132:15133–15135
Bax A, Ikura M (1991) An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the a-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J Biomol NMR 1:99–104
Cady SD et al (2010) Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463:689–692
Castellani F et al (2002) Structure of a protein determined by solid-state magicangle-spinning NMR spectroscopy. Nature 420:98–102
Chevelkov V, Rehbein K, Diel A, Reif B (2006) Ultra-high resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew Chem Int Ed 45:3878–3881
Emsley L, Bodenhausen G (1990) Gaussian pulse cascades: new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem Phys Lett 165:469–476
Franks WT, Kloepper KD, Wylie BJ, Rienstra CM (2007) Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR 39:107–131
Franks WT et al (2008) Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc Natl Acad Sci USA 105:4621–4626
Goddard TD, Kneller DG (2004) SPARKY 3. University of California, San Francisco
Heise H et al (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length a-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA 102:15871–15876
Hiller M et al (2005) Solid-state magic-angle spinning NMR of outer-membrane protein G from Escherichia coli. ChemBioChem 6:1679–1684
Huang KY, Siemer AB, McDermott AE (2011) Homonuclear mixing sequences for perdeuterated proteins. J Magn Reson 208:122–127
Huber M et al (2011) A proton-detected 4D solid-state NMR experiment for protein structure determination. ChemPhysChem 12:915–918
Lange A et al (2006) Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature 440:959–962
Lewandowski JR et al (2011) Enhanced resolution and coherence lifetimes in the solid-state NMR spectroscopy of perdeuterated proteins under ultrafast magic-angle spinning. J Phys Chem Lett 2:2205–2211
Linser R (2011) Side-chain to backbone correlations from solid-state NMR of perdeuterated proteins through combined excitation and long-range magnetization transfers. J Biomol NMR 51:221–226
Linser R, Chevelkov V, Diehl A, Reif B (2007) Sensitivity enhancement using paramagnetic relaxation in MAS solid-state NMR of perdeuterated proteins. J Magn Reson 189:209–216
Linser R, Fink U, Reif B (2008) Proton-detected scalar coupling based assignment strategies in MAS solid-state NMR spectroscopy applied to perdeuterated proteins. J Magn Reson 193:89–93
Linser R, Fink U, Reif B (2010a) Narrow carbonyl resonances in proton-diluted proteins facilitate NMR assignments in the solid state. J Biomol NMR 47:1–6
Linser R, Fink U, Reif B (2010b) Assignment of dynamic regions in biological solids enabled by spin-state selective NMR experiments. J Am Chem Soc 132:8891–8893
Linser R, Bardiaux B, Higman V, Fink U, Reif B (2011a) Structure calculation from unambiguous long-range amide and methyl 1H–1H distance restraints for a micro-crystalline protein with MAS solid state NMR. J Am Chem Soc 133:5905–5912
Linser R et al (2011b) Proton detected solid-state NMR of fibrillar and membrane proteins. Angew Chem Int Ed 50:4508–4512
Loquet A et al (2008) 3D Structure determination of the Crh protein from highly ambiguous solid-state NMR restraints. J Am Chem Soc 130:3579–3589
McDermott AE, Creuzet FJ, Kolbert AC, Griffin RG (1992) High-resolution magic-angle-spinning NMR spectra of protons in deuterated solids. J Magn Reson 98:408–413
Nielsen JT et al (2009) Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy. Angew Chem Int Ed 48:2118–2121
Opella SJ, Marassi FM (2004) Structure determination of membrane proteins by NMR spectroscopy. Chem Rev 104:3587–3606
Paravastu AK, Leapman RD, Yau WM, Tycko R (2008) Molecular structural basis for polymorphism in Alzheimer’s ß-Amyloid fibrils. Proc Natl Acad Sci USA 47:18349–18354
Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H (2001) Backbone and side-chain 13C and 15 N signal assignments of the a-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 Tesla. ChemBioChem 2:272–281
Pervushin KV, Wider G, Wüthrich K (1998) Single transition-to-single transition polarization transfer (ST2-PT) in [15N, 1H]-TROSY. J Biomol NMR 12:345–348
Petkova AT et al (2002) A structural model for Alzheimer’s ß-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA 99:16742–16747
Schanda P, Huber M, Verel R, Ernst M, Meier BH (2009) Direct detection of 3h J NC, hydrogen-bond scalar couplings in proteins by solid-state NMR spectroscopy. Angew Chem Int Ed 48:9322–9325
Schanda P, Meier BH, Ernst M (2010) Quantitative analysis of protein backbone dynamics in microcrystalline ubiquitin by solid-state NMR spectroscopy. J Am Chem Soc 132:15957–15967
Siemer AB et al (2006) Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc 128:13224–13228
Vranken WF et al (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Wasmer C et al (2008) Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science 319:1523–1526
Zhou DH et al (2007a) Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J Am Chem Soc 129:11791–11801
Zhou DH et al (2007b) Solid-state protein structure determination with proton-detected triple resonance 3D magic-angle spinning NMR spectroscopy. Angew Chem Int Ed 46:8380–8383
Acknowledgments
I am grateful to Prof. Bernd Reif for the generous gift of the protein material. Dr. James Hook is kindly acknowledged for his support to the project. This research was financed by the Analytical Centre, UNSW.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Linser, R. Backbone assignment of perdeuterated proteins using long-range H/C-dipolar transfers. J Biomol NMR 52, 151–158 (2012). https://doi.org/10.1007/s10858-011-9593-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-011-9593-2