Abstract
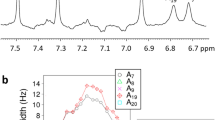
We have analyzed the relaxation properties of all 31P nuclei in an RNA cUUCGg tetraloop model hairpin at proton magnetic field strengths of 300, 600 and 900 MHz in solution. Significant H, P dipolar contributions to R 1 and R 2 relaxation are observed in a protonated RNA sample at 600 MHz. These contributions can be suppressed using a perdeuterated RNA sample. In order to interpret the 31P relaxation data (R 1, R 2), we measured the 31P chemical shift anisotropy (CSA) by solid-state NMR spectroscopy under various salt and hydration conditions. A value of 178.5 ppm for the 31P CSA in the static state (S 2 = 1) could be determined. In order to obtain information about fast time scale dynamics we performed a modelfree analysis on the basis of our relaxation data. The results show that subnanosecond dynamics detected around the phosphodiester backbone are more pronounced than the dynamics detected for the ribofuranosyl and nucleobase moieties of the individual nucleotides (Duchardt and Schwalbe, J Biomol NMR 32:295–308, 2005; Ferner et al., Nucleic Acids Res 36:1928–1940, 2008). Furthermore, the dynamics of the individual phosphate groups seem to be correlated to the 5′ neighbouring nucleobases.




Similar content being viewed by others
References
Akke M, Fiala R, Jiang F, Patel D, Palmer AG 3rd (1997) Base dynamics in a UUCG tetraloop RNA hairpin characterized by 15N spin relaxation: correlations with structure and stability. RNA 3:702–709
Al-Hashimi HM, Walter NG (2008) RNA dynamics: it is about time. Curr Opin Struct Biol 18:321–329
Allain FH, Varani G (1995) Structure of the P1 helix from group I self-splicing introns. J Mol Biol 250:333–353
Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE (2005) Dynamics and metal ion binding in the U6 RNA intramolecular stem-loop as analyzed by NMR. J Mol Biol 353:540–555
Boisbouvier J, Brutscher B, Simorre J, Marion D (1999) 13C spin relaxation measurements in RNA: sensitivity and resolution improvement using spin-state selective correlation experiments. J Biomol NMR 14:241–252
Borer PN, LaPlante SR, Kumar A, Zanatta N, Martin A, Hakkinen A, Levy GC (1994) 13C-NMR relaxation in three DNA oligonucleotide duplexes: model-free analysis of internal and overall motion. Biochemistry 33:2441–2450
Catoire LJ (2004) Phosphorus-31 transverse relaxation rate measurements by NMR spectroscopy: insight into conformational exchange along the nucleic acid backbone. J Biomol NMR 28:179–184
Chowdhury S, Maris C, Allain FH, Narberhaus F (2006) Molecular basis for temperature sensing by an RNA thermometer. EMBO J 25:2487–2497
Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM (1990) Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J Am Chem Soc 112:4989–4991
d’Auvergne EJ, Gooley PR (2003) The use of model selection in the model-free analysis of protein dynamics. J Biomol NMR 25:25–39
Duchardt E, Schwalbe H (2005) Residue specific ribose and nucleobase dynamics of the cUUCGg RNA tetraloop motif by MNMR 13C relaxation. J Biomol NMR 32:295–308
Duchardt E, Nilsson L, Schleucher J (2008) Cytosine ribose flexibility in DNA: a combined NMR 13C spin relaxation and molecular dynamics simulation study. Nucleic Acids Res 36:4211–4219
Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P (2000) The crystal structure of UUCG tetraloop. J Mol Biol 304:35–42
Ferner J, Villa A, Duchardt E, Widjajakusuma E, Wohnert J, Stock G, Schwalbe H (2008) NMR and MD studies of the temperature-dependent dynamics of RNA YNMG-tetraloops. Nucleic Acids Res 36:1928–1940
Ferner J, Suhartono M, Breitung S, Jonker HR, Hennig M, Wohnert J, Gobel M, Schwalbe H (2009) Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries. Chembiochem 10(9):1490–1494
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101
Fürtig B, Buck J, Manoharan V, Bermel W, Jaschke A, Wenter P, Pitsch S, Schwalbe H (2007a) Time-resolved NMR studies of RNA folding. Biopolymers 86:360–383
Fürtig B, Wenter P, Reymond L, Richter C, Pitsch S, Schwalbe H (2007b) Conformational dynamics of bistable RNAs studied by time-resolved NMR spectroscopy. J Am Chem Soc 129:16222–16229
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28:69–79
Fushman D, Tjandra N, Cowburn D (1998) Direct measurement of 15N chemical shift anisotropy in solution. J Am Chem Soc 120:10947–10952
Garcia de la Torre J, Huertas ML, Carrasco B (2000) HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson 147:138–146
Gaudin F, Paquet F, Chanteloup L, Beau JM, Thuong NT, Lancelot G (1995) Selectively C-13-enriched DNA-dynamics of the C1′–H1′ vector in D(CGCAAATTTGCG)(2). J Biomol NMR 5:49–58
Hall JB, Fushman D (2006) Variability of the 15N chemical shielding tensors in the B3 domain of protein G from 15N relaxation measurements at several fields. Implications for backbone order parameters. J Am Chem Soc 128:7855–7870
Hansen AL, Al-Hashimi HM (2006) Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: towards new long-range orientational constraints. J Magn Reson 179:299–307
Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM (2009) Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R(1rho) NMR spectroscopy. J Am Chem Soc 131:3818–3819
Hashim MA-H (2007) Beyond static structures of RNA by NMR: folding, refolding, and dynamics at atomic resolution. Biopolymers 86:345–347
Heisaburo S (1980) NMR relaxation processes of 31P in macromolecules. Biopolymers 19:509–522
Hemminga MA, de Jager PA, Krüse J, Lamerichs RMJN (1987) Magic-angle-spinning NMR on solid biological systems. Analysis of the origin of the spectral linewidths. J Magn Reson 1969(71):446–460
Herzfeld J, Griffin RG, Haberkorn RA (1978) 31P chemical-shift tensors in barium diethyl phosphate and urea-phosphoric acid: model compounds for phospholipid head-group studies. Biochemistry 17:2711–2718
Heus HA, Wijmenga SS, van de Ven FJM, Hilbers CW (1994) Sequential backbone assignment in 13C-Labeled RNA via through-bond coherence transfer using three-dimensional triple resonance spectroscopy (1H, 13C, 31P) and two-dimensional hetero TOCSY. J Am Chem Soc 116:4983–4984
Hobartner C, Micura R (2003) Bistable secondary structures of small RNAs and their structural probing by comparative imino proton NMR spectroscopy. J Mol Biol 325:421–431
Hobartner C, Mittendorfer H, Breuker K, Micura R (2004) Triggering of RNA secondary structures by a functionalized nucleobase. Angew Chem Int Ed Engl 43:3922–3925
Hoogstraten CG, Wank JR, Pardi A (2000) Active site dynamics in the lead-dependent ribozyme. Biochemistry 39:9951–9958
Johnson JE, Hoogstraten CG (2008) Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using (13)C NMR spin relaxation and specific isotope labeling. J Am Chem Soc 130:16757–16769
Johnson JE Jr, Julien KR, Hoogstraten CG (2006) Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J Biomol NMR 35:261–274
Kan JH, Cremers AF, Haasnoot CA, Hilbers CW (1987) The dynamical structure of the RNA in alfalfa mosaic virus studied by 31P-nuclear magnetic resonance. Eur J Biochem 168:635–639
Kellogg GW (1992) Proton-detected hetero-TOCSY experiments with application to nucleic acids. J Magn Reson 1969(98):176–182
Kojima C, Ono A, Kainosho M, James TL (1998) DNA duplex dynamics: NMR relaxation studies of a decamer with uniformly 13C-labeled purine nucleotides. J Magn Reson 135:310–333
Koplin J, Mu Y, Richter C, Schwalbe H, Stock G (2005) Structure and dynamics of an RNA tetraloop: a joint molecular dynamics and NMR study. Structure 13:1255–1267
Kroenke CD, Rance M, Palmer AG (1999) Variability of the 15N chemical shift anisotropy in Escherichia coli ribonuclease H in solution. J Am Chem Soc 121:10119–10125
Lane AN, Jenkins TC, Brown T, Neidle S (1991) Interaction of berenil with the EcoRI dodecamer d(CGCGAATTCGCG)2 in solution studied by NMR. Biochemistry 30:1372–1385
Lang K, Rieder R, Micura R (2007) Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35:5370–5378
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Loth K, Pelupessy P, Bodenhausen G (2005) Chemical shift anisotropy tensors of carbonyl, nitrogen, and amide proton nuclei in proteins through cross-correlated relaxation in NMR spectroscopy. J Am Chem Soc 127:6062–6068
Lynch SR, Puglisi JD (2001) Structure of a eukaryotic decoding region A-site RNA. J Mol Biol 306:1023–1035
Magusin PC, Hemminga MA (1993) A theoretical study of rotational diffusion models for rod-shaped viruses. The influence of motion on 31P nuclear magnetic resonance lineshapes and transversal relaxation. Biophys J 64:1851–1860
Magusin PC, Hemminga MA (1994) Analysis of 31P MAS NMR spectra and transversal relaxation of bacteriophage M13 and tobacco mosaic virus. Biophys J 66:1197–1208
Mandel AM, Akke M, Palmer IIIAG (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol 246:144–163
Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C (1994) Three-dimensional triple-resonance 1H, 13C, 31P experiment: sequential through-bond correlation of ribose protons and intervening phosphorus along the RNA oligonucleotide backbone. J Am Chem Soc 116:6472–6473
Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C (1995) Sequential correlation of anomeric ribose protons and intervening phosphorus in RNA oligonucleotides by a 1H, 13C, 31P triple resonance experiment: HCP-CCH-TOCSY. J Biomol NMR 5:87–92
Miller JL, Kollman PA (1997) Theoretical studies of an exceptionally stable RNA tetraloop: observation of convergence from an incorrect NMR structure to the correct one using unrestrained molecular dynamics. J Mol Biol 270:436–450
Narberhaus F, Waldminghaus T, Chowdhury S (2006) RNA thermometers. FEMS Microbiol Rev 30:3–16
Nina M, Simonson T (2002) Molecular dynamics of the tRNAAla acceptor stem: comparison between continuum reaction field and particle-mesh Ewald electrostatic treatments. J Phys Chem B 106:3696–3705
Oberstrass FC, Allain FH, Ravindranathan S (2008) Changes in dynamics of SRE-RNA on binding to the VTS1p-SAM domain studied by 13C NMR relaxation. J Am Chem Soc 130:12007–12020
Odahara T, Nishimoto S, Katsutani N, Kyogoku Y, Morimoto Y, Matsushiro A, Akutsu H (1994) Dynamic properties of nucleic acids in biosupramolecular systems, as studied by 31P NMR. J Biochem 115:270–278
Olsen JI, Schweizer MP, Walkiw IJ, Hamill WD Jr, Horton WJ, Grant DM (1982) Carbon-13 NMR relaxation studies of pre-melt structural dynamics in [4–13C-uracil] labeled E. coli transfer RNAIVal. Nucleic Acids Res 10:4449–4464
Precechtelova J, Padrta P, Munzarova ML, Sklenar V (2008) 31P chemical shift tensors for canonical and non-canonical conformations of nucleic acids: a DFT study and NMR implications. J Phys Chem B 112:3470–3478
Reichert D, Pascui O, deAzevedo ER, Bonagamba TJ, Arnold K, Huster D (2004) A solid-state NMR study of the fast and slow dynamics of collagen fibrils at varying hydration levels. Magn Reson Chem 42:276–284
Rieder R, Lang K, Graber D, Micura R (2007) Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem 8:896–902
Schmidt PG, Playl T, Agris PF (1983) Internal dynamics of transfer ribonucleic acid determined by nuclear magnetic resonance of carbon-13-enriched ribose carbon 1. Biochemistry 22:1408–1415
Schmidt PG, Sierzputowska-Gracz H, Agris PF (1987) Internal motions in yeast phenylalanine transfer RNA from 13C NMR relaxation rates of modified base methyl groups: a model-free approach. Biochemistry 26:8529–8534
Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J (2007) Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl 46:1212–1219
Shajani Z, Varani G (2005) 13C NMR relaxation studies of RNA base and ribose nuclei reveal a complex pattern of motions in the RNA binding site for human U1A protein. J Mol Biol 349:699–715
Shajani Z, Varani G (2007) NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers 86:348–359
Shajani Z, Drobny G, Varani G (2007) Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry 46:5875–5883
Showalter SA, Baker NA, Tang C, Hall KB (2005) Iron responsive element RNA flexibility described by NMR and isotropic reorientational eigenmode dynamics. J Biomol NMR 32:179–193
Spielmann HP (1998) Dynamics in psoralen-damaged DNA by 1H-detected natural abundance 13C NMR spectroscopy. Biochemistry 37:5426–5438
Terao T, Matsui S, Akasaka K (1977) Phosphorus-31 chemical shift anisotropy in solid nucleic acids. J Am Chem Soc 99:6136–6138
Tucker BJ, Breaker RR (2005) Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15:342–348
Vallurupalli P, Kay LE (2005) A suite of 2H NMR spin relaxation experiments for the measurement of RNA dynamics. J Am Chem Soc 127:6893–6901
Varani G, Tinoco I Jr (1991) RNA structure and NMR spectroscopy. Q Rev Biophys 24:479–532
Villa A, Stock G (2006) What NMR relaxation can tell us about the internal motion of an RNA Hairpin: a molecular dynamics simulation study. J. Chem. Theory Comput. 2:1228–1236
Villa A, Widjajakusuma E, Stock G (2008) Molecular dynamics simulation of the structure, dynamics, and thermostability of the RNA hairpins uCACGg and cUUCGg. J Phys Chem B 112:134–142
Vokacova Z, Budesinsky M, Rosenberg I, Schneider B, Sponer J, Sychrovsky V (2009) Structure and dynamics of the ApA, ApC, CpA, and CpC RNA dinucleoside monophosphates resolved with NMR scalar spin–spin couplings. J Phys Chem B 113:1182–1191
Waldminghaus T, Heidrich N, Brantl S, Narberhaus F (2007) FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 65:413–424
Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S (2005) Kinetics of photoinduced RNA refolding by real-time NMR spectroscopy. Angew Chem Int Ed Engl 44:2600–2603
Wenter P, Bodenhausen G, Dittmer J, Pitsch S (2006a) Kinetics of RNA refolding in dynamic equilibrium by 1H-detected 15N exchange NMR spectroscopy. J Am Chem Soc 128:7579–7587
Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S (2006b) A caged uridine for the selective preparation of an RNA fold and determination of its refolding kinetics by real-time NMR. Chembiochem 7:417–420
Williams DJ, Hall KB (1999) Unrestrained stochastic dynamics simulations of the UUCG tetraloop using an implicit solvation model. Biophys J 76:3192–3205
Williams DJ, Hall KB (2000) Experimental and theoretical studies of the effects of deoxyribose substitutions on the stability of the UUCG tetraloop. J Mol Biol 297:251–265
Williams DJ, Boots JL, Hall KB (2001) Thermodynamics of 2′-ribose substitutions in UUCG tetraloops. RNA 7:44–53
Williamson JR, Boxer SG (1989a) Multinuclear NMR studies of DNA hairpins. 1. Structure and dynamics of d(CGCGTTGTTCGCG). Biochemistry 28:2819–2831
Williamson JR, Boxer SG (1989b) Multinuclear NMR studies of DNA hairpins. 2. Sequence-dependent structural variations. Biochemistry 28:2831–2836
Winkler WC (2005) Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr Opin Chem Biol 9:594–602
Zhang Q, Sun X, Watt ED, Al-Hashimi HM (2006) Resolving the motional modes that code for RNA adaptation. Science 311:653–656
Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM (2007) Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450:1263–1267
Acknowledgments
The work has been supported by the state of Hesse (Center for Biomolecular Magnetic Resonance, BMRZ) and the DFG (Sonderforschungsbereich 579: RNA-ligand-interactions). H·S. and C.G. are members of the DFG Cluster of Excellence: Macromolecular Complexes (EXC 115).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rinnenthal, J., Richter, C., Nozinovic, S. et al. RNA phosphodiester backbone dynamics of a perdeuterated cUUCGg tetraloop RNA from phosphorus-31 NMR relaxation analysis. J Biomol NMR 45, 143–155 (2009). https://doi.org/10.1007/s10858-009-9343-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9343-x