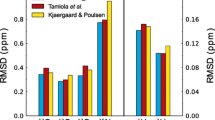
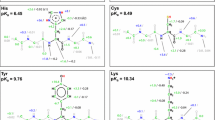
Abstract
We propose a new computational model to predict amide proton chemical shifts in proteins. In addition to the ring-current, susceptibility and electrostatic effects of earlier models, we add a hydrogen-bonding term based on density functional calculations of model peptide–peptide and peptide–water systems. Both distance and angular terms are included, and the results are rationalized in terms of natural bond orbital analysis of the interactions. Comparison to observed shifts for 15 proteins shows a significant improvement over existing structure-shift correlations. These additions are incorporated in a new version of the SHIFTS program.











Similar content being viewed by others
References
Asakura T, Taoka K, Demura M, Williamson MP (1995) The relationship between amide proton chemical shifts and secondary structure. J Biomol NMR 6:227–236
Barfield M (2002) Structural dependencies of interresidue scalar coupling h3JNC’ and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J Am Chem Soc 124:4158–4168
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Beger RD, Bolton PH (1997) Protein ϕ and ψ dihedral restraints determined from multidimensional hypersurface correlations of backbone chemical shifts and their use in the determination of protein tertiary structures. J Biomol NMR 10:129–142
Bohmann JA, Weinhold F, Farrar TC (1997) Natural chemical shielding analysis of nuclear magnetic resonance shielding tensors from gauge-including atomic orbital calculations. J Chem Phys 107:1173–1184
Buckingham AD, Schaefer T, Schneider WG (1960) Solvent effects in nuclear magnetic resonance spectra. J Chem Phys 32:1227–1233
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods R (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
Cui Q, Karplus M (2000) Molecular properties from combined QM/MM methods 2. Chemical shifts in large molecules. J Phys Chem B 104:3721–3743
de Dios AC, Pearson JG, Oldfield E (1993) Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science 260:1491–1496
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Frisch MJ, Trucks GW, Schlegel HB, Gill PMW, Johnson BG, Robb JA, Cheeseman JR, Keith TA, Petersson GA, Montgomery JA, Raghavachari K, Al-Laham MA, Zakrzewski VG, Ortiz JV, Foresman JB, Cioslowski J, Stefanov BB, Nanayakkara A, Challacombe M, Peng CY, Ayala PY, Chen W, Wong MW, Andres JL, Replogle ES, Gomperts R, Martin RL, Fox DJ, Binkley JS, Defrees DJ, Baker J, Stewart JP, Head-Gordon M, Gonzalez C, Pople JA (1998) Gaussian 98 (Revision A9). Gaussian Inc, Pittsburgh PA
Haigh CW, Mallion RB (1980) Ring current theories in nuclear magnetic resonance. Prog NMR Spectr 13:303–344
Iwadate M, Asakura T, Williamson MP (1999) Cα and Cβ carbon-13 chemical shifts in proteins from an empirical database. J Biomol NMR 13:199–211
Jeng M-F, Campbell AP, Begley T, Holmgren A, Case DA, Wright PE, Dyson HJ (1994) High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 2:853–868
Jorgensen WL, Chandrasekhar J, Madura J, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Le H, Oldfield E (1994) Correlation between N NMR chemical shifts in proteins and secondary structure. J Biomol NMR 4:341–348
Le H, Oldfield E (1996) Ab initio studies of amide-N chemical shifts in dipeptides: applications to protein NMR spectroscopy. J Phys Chem 100:16423–16428
McConnell HM (1957) Theory of nuclear magnetic shielding in molecules I Long-range dipolar shielding of protons. J Chem Phys 27:226–229
Meiler J (2003) PROSHIFT: protein chemical shift prediction using artificial neural networks. J Biomol NMR 26:25–37
Meiler J, Maier W, Will M, Meusinger R (2002) Using neural networks for 13C NMR chemical shift prediction-comparison with traditional methods. J Magn Reson 157:242–252
Moon S, Case DA (2006) A comparison of quantum chemical models for calculating NMR shielding parameters in peptides: mixed basis set and ONIOM methods combined with a complete basis set extrapolation. J Comput Chem 27:825–836
Neal S, Nip AM, Zhang H, Wishart DS (2003) Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR 26:215–240
Ösapay K, Case DA (1991) A new analysis of proton chemical shifts in proteins. J Am Chem Soc 113:9436–9444
Parker LL, Houk AR, Jensen JH (2006) Cooperative hydrogen bonding effects are key determinants of backbone amide proton chemical shifts in proteins. J Am Chem Soc 128:9863–9872
Pearson JG, Le H, Sanders LK, Godbout N, Havlin RH, Oldfield E (1997) Predicting chemical shifts in proteins: structure refinement of valine residues by using ab initio and empirical geometry optimizations. J Am Chem Soc 119:11941–11950
Perdew JP, Wang Y (1992) Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B 45:13244–13249
Polshakov VI, Birdsall B, Feeney J (1999) Characterization of rates of ring-flipping in trimethoprim in its ternary complexes with Lactobacillus casei dihydrofolate reductase and coenzyme analogues. Biochemistry 38:15962–15969
Redfield C, Dobson CM (1990) H NMR studies of human lysozyme: spectral assignment and comparison with hen lysozyme. Biochemistry 29:7201–7214
Rumelhart DE, McClelland J (1986) Parallel distrbuted processing. MIT Press, Boston
Sharma Y, Kwon OY, Brooks B, Tjandra N (2002) An ab initio study of amide proton shift tensor dependence on local protein structure. J Am Chem Soc 124:327–335
Sitkoff D, Case DA (1997) Density functional calculations of proton chemical shifts in model peptides. J Am Chem Soc 119:12262–12273
Spera S, Bax A (1991) Empirical correlation between protein backbone conformation and Cα and Cβ C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113:5490–5492
Wishart DS, Case DA (2001) Use of chemical shifts in macromolecular structure determination. Meth Enzymol 338:3–34
Xu XP, Case DA (2001) Automated prediction of N, Cα, Cβ and C’ chemical shifts in proteins using a density functional database. J Biomol NMR 21:321–333
Xu XP, Case DA (2002) Probing multiple effects on N, Cα, Cβ and C’ chemical shifts in peptides using density functional theory. Biopolymers 65:408–423
Zupan J, Gasteiger J (1993) Neural networks for chemists. VCH Verlagsgesellschaft mbH, Weinheim
Acknowledgments
This work was supported by NIH grant GM45811. We thank Jan Ziegler, Stephan Schwarzinger and Jan Jensen for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Moon, S., Case, D.A. A new model for chemical shifts of amide hydrogens in proteins. J Biomol NMR 38, 139–150 (2007). https://doi.org/10.1007/s10858-007-9156-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9156-8