Abstract
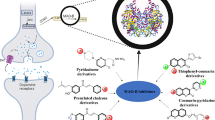
The c-Jun N-terminal kinase 3 (JNK3) signaling cascade is activated during cerebral ischemia leading to neuronal damage. The present study was carried out to identify and evaluate novel JNK3 inhibitors using in-silico and in-vitro approach. A total of 380 JNK3 inhibitors belonging to different organic groups was collected from the previously reported literature. These molecules were used to generate a pharmacophore model. This model was used to screen a chemical database (SPECS) to identify newer molecules with similar chemical features. The top 1000 hits molecules were then docked against the JNK3 enzyme coordinate following GLIDE rigid receptor docking (RRD) protocol. Best posed molecules of RRD were used during induced-fit docking (IFD), allowing receptor flexibility. Other computational predictions such as binding free energy, electronic configuration and ADME/tox were also calculated. Inferences from the best pharmacophore model suggested that, in order to have specific JNK3 inhibitory activity, the molecules must possess one H-bond donor, two hydrophobic and two ring features. Docking studies suggested that the main interaction between lead molecules and JNK3 enzyme consisted of hydrogen bond interaction with methionine 149 of the hinge region. It was also observed that the molecule with better MM-GBSA dG binding free energy, had greater correlation with JNK3 inhibition. Lead molecule (AJ-292-42151532) with the highest binding free energy (dG = 106.8 Kcal/mol) showed better efficacy than the SP600125 (reference JNK3 inhibitor) during cell-free JNK3 kinase assay (IC50 = 58.17 nM) and cell-based neuroprotective assay (EC50 = 7.5 µM).
Graphic Abstract








Similar content being viewed by others
References
Pandian JD, Sudhan P (2013) Stroke epidemiology and stroke care services in India. J Stroke 15:128. https://doi.org/10.5853/jos.2013.15.3.128
Feigin VL, Krishnamurthi RV, Parmar P et al (2015) Update on the global burden of ischemic and hemorrhagic stroke in 1990–2013: the GBD 2013 Study. Neuroepidemiology 45:161–176. https://doi.org/10.1159/000441085
Shao S, Xu M, Zhou J et al (2017) Atorvastatin attenuates ischemia/reperfusion-induced hippocampal neurons injury via Akt-nNOS-JNK signaling pathway. Cell Mol Neurobiol 37:753–762. https://doi.org/10.1007/s10571-016-0412-x
Kuan C-Y, Whitmarsh AJ, Yang DD et al (2003) A critical role of neural-specific JNK3 for ischemic apoptosis. Proc Natl Acad Sci 100:15184–15189. https://doi.org/10.1073/pnas.2336254100
Messoussi A, Feneyrolles C, Bros A et al (2014) Recent progress in the design, study, and development of c-Jun N-terminal kinase inhibitors as anticancer agents. Chem Biol 21:1433–1443. https://doi.org/10.1016/j.chembiol.2014.09.007
Wu X-X, Dai D-S, Zhu X et al (2014) Molecular modeling studies of JNK3 inhibitors using QSAR and docking. Med Chem Res 23:2456–2475. https://doi.org/10.1007/s00044-013-0782-2
Xu P, Yoshioka K, Yoshimura D et al (2003) In vitro development of mouse embryonic stem cells lacking JNK/stress-activated protein kinase-associated protein 1 (JSAP1) scaffold protein revealed its requirement during early embryonic neurogenesis. J Biol Chem 278:48422–48433. https://doi.org/10.1074/jbc.M307888200
Liu J-R, Zhao Y, Patzer A et al (2010) The c-Jun N-terminal kinase (JNK) inhibitor XG-102 enhances the neuroprotection of hyperbaric oxygen after cerebral ischaemia in adult rats. Neuropathol Appl Neurobiol 36:211–224. https://doi.org/10.1111/j.1365-2990.2009.01047.x
Jung H, Aman W, Hah J-M (2017) Novel scaffold evolution through combinatorial 3D-QSAR model studies of two types of JNK3 inhibitors. Bioorg Med Chem Lett 27:2139–2143. https://doi.org/10.1016/j.bmcl.2017.03.063
Zhang ZB, Li ZG (2012) Cathepsin B and Phospo-JNK in relation to ongoing apoptosis after transient focal cerebral ischemia in the rat. Neurochem Res 37:948–957. https://doi.org/10.1007/s11064-011-0687-8
Yamasaki T, Kawasaki H, Nishina H (2012) Diverse roles of JNK and MKK pathways in the brain. J Signal Transduct 2012:1–9. https://doi.org/10.1155/2012/459265
Yarza R, Vela S, Solas M, Ramirez MJ (2016) c-Jun N-terminal kinase (JNK) signaling as a therapeutic target for Alzheimer’s disease. Front Pharmacol.https://doi.org/10.3389/fphar.2015.00321
Mishra P, Günther S (2018) New insights into the structural dynamics of the kinase JNK3. Sci Rep 8:9435. https://doi.org/10.1038/s41598-018-27867-3
Xie X, Gu Y, Fox T et al (1998) Crystal structure of JNK3: a kinase implicated in neuronal apoptosis. Structure 6:983–991. https://doi.org/10.1016/S0969-2126(98)00100-2
Assi K, Pillai R, Gomez-Munoz A et al (2006) The specific JNK inhibitor SP600125 targets tumour necrosis factor-alpha production and epithelial cell apoptosis in acute murine colitis. Immunology 118:112–121. https://doi.org/10.1111/j.1365-2567.2006.02349.x
Gross ND, Boyle JO, Du B et al (2007) Inhibition of Jun NH2-terminal kinases suppresses the growth of experimental head and neck squamous cell carcinoma. Clin Cancer Res 13:5910–5917. https://doi.org/10.1158/1078-0432.CCR-07-0352
Minutoli L, Altavilla D, Marini H et al (2004) Protective effects of SP600125 a new inhibitor of c-jun N-terminal kinase (JNK) and extracellular-regulated kinase (ERK1/2) in an experimental model of cerulein-induced pancreatitis. Life Sci 75:2853–2866. https://doi.org/10.1016/j.lfs.2004.03.040
Gaillard P, Jeanclaude-Etter I, Ardissone V et al (2005) Design and synthesis of the first generation of novel potent, selective, and in vivo active (Benzothiazol-2-yl)acetonitrile inhibitors of the c-Jun N-terminal kinase. J Med Chem 48:4596–4607. https://doi.org/10.1021/jm0310986
Rückle T, Biamonte M, Grippi-Vallotton T et al (2004) Design, synthesis, and biological activity of novel, potent, and selective (Benzoylaminomethyl)thiophene sulfonamide inhibitors of c-Jun-N-terminal kinase. J Med Chem 47:6921–6934. https://doi.org/10.1021/jm031112e
Swahn B-M, Huerta F, Kallin E et al (2005) Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3. Bioorg Med Chem Lett 15:5095–5099. https://doi.org/10.1016/j.bmcl.2005.06.083
Swahn B-M, Xue Y, Arzel E et al (2006) Design and synthesis of 2′-anilino-4,4′-bipyridines as selective inhibitors of c-Jun N-terminal kinase-3. Bioorg Med Chem Lett 16:1397–1401. https://doi.org/10.1016/j.bmcl.2005.11.039
Cao J, Gao H, Bemis G et al (2009) Structure-based design and parallel synthesis of N-benzyl isatin oximes as JNK3 MAP kinase inhibitors. Bioorg Med Chem Lett 19:2891–2895. https://doi.org/10.1016/j.bmcl.2009.03.043
Shin Y, Chen W, Habel J et al (2009) Synthesis and SAR of piperazine amides as novel c-jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 19:3344–3347. https://doi.org/10.1016/j.bmcl.2009.03.086
Song X, Chen W, Lin L et al (2011) Synthesis and SAR of 2-phenoxypyridines as novel c-Jun N-terminal kinase inhibitors. Bioorg Med Chem Lett 21:7072–7075. https://doi.org/10.1016/j.bmcl.2011.09.090
Zheng K, Iqbal S, Hernandez P et al (2014) Design and synthesis of highly potent and isoform selective JNK3 Inhibitors: SAR studies on aminopyrazole derivatives. J Med Chem 57:10013–10030. https://doi.org/10.1021/jm501256y
He Y, Duckett D, Chen W et al (2014) Synthesis and SAR of novel isoxazoles as potent c-jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 24:161–164. https://doi.org/10.1016/j.bmcl.2013.11.052
Zheng K, Park CM, Iqbal S et al (2015) Pyridopyrimidinone derivatives as potent and selective c-Jun N-terminal kinase (JNK) inhibitors. ACS Med Chem Lett 6:413–418. https://doi.org/10.1021/ml500474d
Darshit BS, Balaji B, Rani P, Ramanathan M (2014) Identification and in vitro evaluation of new leads as selective and competitive glycogen synthase kinase-3β inhibitors through ligand and structure based drug design. J Mol Graph Model 53:31–47. https://doi.org/10.1016/j.jmgm.2014.06.013
Dixon SL, Smondyrev AM, Knoll EH et al (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. https://doi.org/10.1007/s10822-006-9087-6
Muralikumar S, Vetrivel U, Narayanasamy A, Das N U (2017) Probing the intermolecular interactions of PPARγ-LBD with polyunsaturated fatty acids and their anti-inflammatory metabolites to infer most potential binding moieties. Lipids Health Dis 16:17. https://doi.org/10.1186/s12944-016-0404-3
Hevener KE, Mehboob S, Su P-C et al (2012) Discovery of a novel and potent class of F. tularensis Enoyl-Reductase (FabI) inhibitors by molecular shape and electrostatic matching. J Med Chem 55:268–279. https://doi.org/10.1021/jm201168g
Jana S, Singh SK (2019) Identification of selective MMP-9 inhibitors through multiple e-pharmacophore, ligand-based pharmacophore, molecular docking, and density functional theory approaches. J Biomol Struct Dyn 37:944–965. https://doi.org/10.1080/07391102.2018.1444510
Sivashanmugam M, S KN (2019) Virtual screening of natural inhibitors targeting ornithine decarboxylase with pharmacophore scaffolding of DFMO and validation by molecular dynamics simulation studies. J Biomol Struct Dyn 37:766–780. https://doi.org/10.1080/07391102.2018.1439772
Zhang G, Musgrave CB (2007) Comparison of DFT methods for molecular orbital eigenvalue calculations. J Phys Chem A 111:1554–1561. https://doi.org/10.1021/jp061633o
Pires DEV, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Yang H, Lou C, Sun L et al (2019) admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 35:1067–1069. https://doi.org/10.1093/bioinformatics/bty707
Darshit BS, Ramanathan M (2016) Activation of AKT1/GSK-3β/β-catenin–TRIM11/survivin pathway by novel GSK-3β inhibitor promotes neuron cell survival: study in differentiated SH-SY5Y cells in OGD model. Mol Neurobiol 53:6716–6729. https://doi.org/10.1007/s12035-015-9598-z
Wu X (2015) Dual AO/EB staining to detect apoptosis in osteosarcoma cells compared with flow cytometry. Med Sci Monit Basic Res 21:15–20. https://doi.org/10.12659/MSMBR.893327
Banavath HN, Sharma OP, Kumar MS, Baskaran R (2015) Identification of novel tyrosine kinase inhibitors for drug resistant T315I mutant BCR-ABL: a virtual screening and molecular dynamics simulations study. Sci Rep 4:6948. https://doi.org/10.1038/srep06948
Manolopoulos DE, May C, Down SE (1991) Theoretical studies of the fullerenes: CJ4 to CT0. Chem Phys Lett 181:105–111
Acknowledgements
We thank Shelvia Malik, Jr. Application scientist, Dr. Sudharsan Pandiyan, Senior Application Scientist, Schrodinger, and Dr. Pritesh Bhat, Senior Application Scientist, Schrodinger, for providing their valuable time and expertise and technical support during the study. We thank the National Centre for Cell Science (NCCS) Pune for providing cell lines at a reasonable price and in a timely manner. We also thank, Dr. Sivaram Hariharan for syntax and grammar revision. We thank PSG Sons & Charity and Tamilnadu Dr. MGR medical university for providing us with a better facility and environment for the learning.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rajan, R.K., Ramanathan, M. Identification and neuroprotective evaluation of a potential c-Jun N-terminal kinase 3 inhibitor through structure-based virtual screening and in-vitro assay. J Comput Aided Mol Des 34, 671–682 (2020). https://doi.org/10.1007/s10822-020-00297-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00297-y