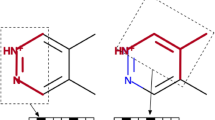
Abstract
A variety of fields would benefit from accurate \(pK_a\) predictions, especially drug design due to the effect a change in ionization state can have on a molecule’s physiochemical properties. Participants in the recent SAMPL6 blind challenge were asked to submit predictions for microscopic and macroscopic \(pK_a\)s of 24 drug like small molecules. We recently built a general model for predicting \(pK_a\)s using a Gaussian process regression trained using physical and chemical features of each ionizable group. Our pipeline takes a molecular graph and uses the OpenEye Toolkits to calculate features describing the removal of a proton. These features are fed into a Scikit-learn Gaussian process to predict microscopic \(pK_a\)s which are then used to analytically determine macroscopic \(pK_a\)s. Our Gaussian process is trained on a set of 2700 macroscopic \(pK_a\)s from monoprotic and select diprotic molecules. Here, we share our results for microscopic and macroscopic predictions in the SAMPL6 challenge. Overall, we ranked in the middle of the pack compared to other participants, but our fairly good agreement with experiment is still promising considering the challenge molecules are chemically diverse and often polyprotic while our training set is predominately monoprotic. Of particular importance to us when building this model was to include an uncertainty estimate based on the chemistry of the molecule that would reflect the likely accuracy of our prediction. Our model reports large uncertainties for the molecules that appear to have chemistry outside our domain of applicability, along with good agreement in quantile–quantile plots, indicating it can predict its own accuracy. The challenge highlighted a variety of means to improve our model, including adding more polyprotic molecules to our training set and more carefully considering what functional groups we do or do not identify as ionizable.









Similar content being viewed by others
References
Wan H, Ulander J (2006) High-throughput pKa screening and prediction amenable for ADME profiling. Expert Opin Drug Metab Toxicol 2(1):139. https://doi.org/10.1517/17425255.2.1.139
Gleeson MP (2008) Generation of a set of simple, interpretable ADMET rules of thumb. J Med Chem 51(4):817. https://doi.org/10.1021/jm701122q
Manallack DT, Prankerd RJ, Yuriev E, Oprea TI, Chalmers DK (2013) The significance of acid/base properties in drug discovery. Chem Soc Rev 42(2):485. https://doi.org/10.1039/c2cs35348b
Manchester J, Walkup G, Rivin O, You Z (2010) Evaluation of pKa estimation methods on 211 druglike compounds. J Chem Inf Model 50(4):565. https://doi.org/10.1021/ci100019p
Settimo L, Bellman K, Knegtel RMA (2014) Comparison of the accuracy of experimental and predicted pKa values of basic and acidic compounds. Pharm Res 31(4):1082. https://doi.org/10.1007/s11095-013-1232-z
Fraczkiewicz R (2013) In silico prediction of ionization. In: Reedijk J (ed) Reference module in chemistry, molecular sciences and chemical engineering. Elsevier, Waltham
Bannan CC, Burley KH, Chiu M, Shirts MR, Gilson MK, Mobley DL (2016) Blind prediction of cyclohexane–water distribution coefficients from the SAMPL5 challenge. J Comput Aided Mol Des 30(11):1. https://doi.org/10.1007/s10822-016-9954-8
Pickard FC, König G, Tofoleanu F, Lee J, Simmonett AC, Shao Y, Ponder JW, Brooks BR (2016) Blind prediction of distribution in the SAMPL5 challenge with QM based protomer and pKa corrections. J Comput Aided Mol Des 30(11):1. https://doi.org/10.1007/s10822-016-9955-7
Aguilar B, Anandakrishnan R, Ruscio JZ, Onufriev AV (2010) Statistics and physical origins of pK and ionization state changes upon protein-ligand binding. Biophys J 98(5):872. https://doi.org/10.1016/j.bpj.2009.11.016
Işık M, Levorse D, Rustenburg AS, Ndukwe IE, Wang H, Wang X, Reibarkh M, Martin GE, Makarov AA, Mobley DL, Rhodes T, Chodera JD (2018) pka measurements for the sampl6 prediction challenge for a set of kinase inhibitor-like fragments. bioRxiv. https://doi.org/10.1101/368787. https://www.biorxiv.org/content/early/2018/07/13/368787
Darvey IG (1995) The assignment of pKa values to functional groups in amino acids. Biochem Educ 23(2):80. https://doi.org/10.1016/0307-4412(94)00150-N
Bodner GM (1986) Assigning the pKa’s of polyprotic acids. J Chem Educ 63(3):246. https://doi.org/10.1021/ed063p246
Işık M, Rustenburg AS (2018) Michael, Shirts, D.L. Mobley, J.D. Chodera. SAMPL6. https://github.com/MobleyLab/SAMPL6
Exner O (1972) Advances in linear free energy relationships. Springer, Boston. https://doi.org/10.1007/978-1-4615-8660-9_1
Perrin D, Dempsey B, Serjeant E (1981) pKa prediction for organic acids and bases. Chapman and Hall, New York
Geidl S, Svobodová Vařeková R, Bendová V, Petrusek L, Ionescu CM, Jurka Z, Abagyan R, Koča J (2015) How does the methodology of 3D structure preparation influence the quality of pKa prediction? J Chem Inf Model 55(6):1088. https://doi.org/10.1021/ci500758w
Cruciani G, Milletti F, Storchi L, Sforna G, Goracci L (2009) In silico pKa prediction and ADME profiling. Chem Biodivers. 6(11):1812. https://doi.org/10.1002/cbdv.200900153
Katritzky AR, Kuanar M, Slavov S, Hall CD, Karelson M, Kahn I, Dobchev DA (2010) Quantitative correlation of physical and chemical properties with chemical structure: utility for prediction. Chem Rev 110(10):5714. https://doi.org/10.1021/cr900238d
Peterson KL (2000) Reviews in computational chemistry. Wiley, Hoboken
Fraczkiewicz R, Lobell M, Göller AH, Krenz U, Schoenneis R, Clark RD, Hillisch A (2015) Best of both worlds: combining pharma data and state of the art modeling technology to improve in silico pKa prediction. J Chem Inf Model 55(2):389. https://doi.org/10.1021/ci500585w
Citra MJ (1999) Estimating the pKa of phenols, carboxylic acids and alcohols from semi-empirical quantum chemical methods. Chemosphere 38(1):191. https://doi.org/10.1016/S0045-6535(98)00172-6
Vařeková RS, Geidl S, Ionescu CM, Skřehota O, Bouchal T, Sehnal D, Abagyan R, Koča J (2013) Predicting pKa values from EEM atomic charges. J Cheminf 5:18. https://doi.org/10.1186/1758-2946-5-18
Dixon SL, Jurs PC (1993) Estimation of pKa for organic oxyacids using calculated atomic charges. J Comput Chem 14(12):1460. https://doi.org/10.1002/jcc.540141208
Zevatskii YE, Samoilov DV (2011) Modern methods for estimation of ionization constants of organic compounds in solution. Russ J Org Chem 47(10):1445. https://doi.org/10.1134/S1070428011100010
Pracht P, Bauer CA, Grimme S (2017) Automated and efficient quantum chemical determination and energetic ranking of molecular protonation sites. J Comput Chem 38(30):2618. https://doi.org/10.1002/jcc.24922
Bochevarov AD, Harder E, Hughes TF, Greenwood JR, Braden DA, Philipp DM, Rinaldo D, Halls MD, Zhang J, Friesner RA, Jaguar (2013) A high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem 113(18):2110. https://doi.org/10.1002/qua.24481
Bochevarov AD, Watson MA, Greenwood JR, Philipp DM (2016) Multiconformation, density functional theory-based pka prediction in application to large, flexible organic molecules with diverse functional groups. J Chem Theory Comput 12(12):6001. https://doi.org/10.1021/acs.jctc.6b00805
Rasmussen CE, Williams CKI (2006) Gaussian processes for machine learning, adaptive computation and machine learning. MIT Press, Cambridge
OpeneEye Scientific Software, Inc. OEChem Toolkit (2018). http://www.eyesopen.com
Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and cambridge structural database. J Chem Inf Model 50(4):572. https://doi.org/10.1021/ci100031x
Wiberg KB (1968) Application of the pople-santry-segal CNDO method to the cyclopropylcarbinyl and cyclobutyl cation and to bicyclobutane. Tetrahedron 24(3):1083. https://doi.org/10.1016/0040-4020(68)88057-3
Mayer I (2007) Bond order and valence indices: a personal account. J Comput Chem 28(1):204. https://doi.org/10.1002/jcc.20494
OpeneEye Scientific Software, Inc. OEQuacPac Toolkit (2018). http://www.eyesopen.com
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J Comput Chem 21(2):132
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23(16):1623. https://doi.org/10.1002/jcc.10128
Jelfs S, Ertl P, Selzer P (2007) Estimation of pKa for druglike compounds using semiempirical and information-based descriptors. J Chem Inf Model 47(2):450. https://doi.org/10.1021/ci600285n
Nicholls A, Wlodek S, Grant JA (2010) SAMPL2 and continuum modeling. J Comput Aided Mol Des 24(4):293. https://doi.org/10.1007/s10822-010-9334-8
Grant JA, Pickup BT, Nicholls A (2001) A smooth permittivity function for Poisson-Boltzmann solvation methods. J Comput Chem 22(6):608. https://doi.org/10.1002/jcc.1032
Nicholls A (2004) Spicoli: a surface toolkit, dude
Lee B, Richards FM (1971) The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55(3):379. https://doi.org/10.1016/0022-2836(71)90324-X
Connolly ML (1983) Analytical molecular surface calculation. J Appl Cryst 16(5):548. https://doi.org/10.1107/S0021889883010985
Sharp KA, Nicholls A, Fine RF, Honig B (1991) Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science 252(5002):106. https://doi.org/10.1126/science.2011744
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825
Kortüm G, Vogel W, Andrussow K (1960) Disssociation constants of organic acids in aqueous solution. Pure Appl Chem 1(2–3):187. https://doi.org/10.1351/pac196001020187
Perrin DD (1972) Dissociation constants of organic bases in aqueous solution: supplement 1972. Butterworths, London
Serjeant P, Dempsey B (1979) Ionisation constants of organic acids in aqueous solution. Pergamon, Oxford
Hastie T, Tibshirani R, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction, 2nd edn. Springer, New York
Kuhn HW (2004) The Hungarian method for the assignment problem. Nav Res Logist 52(1):7. https://doi.org/10.1002/nav.20053
Advanced Chemistry Development, Inc. pKa GALAS (2015). www.acdlabs.com
Ripin D, Evans D (2005) pKa table. http://evans.rc.fas.harvard.edu/pdf/evans_pKa_table.pdf
Goldfarb AR, Mele A, Gutstein N (1955) Basicity of the amide bond. J Am Chem Soc 77(23):6194. https://doi.org/10.1021/ja01628a031
Bordwell FG, Algrim DJ, Harrelson JA (1988) The relative ease of removing a proton, a hydrogen atom, or an electron from carboxamides versus thiocarboxamides. J Am Chem Soc 110(17):5903. https://doi.org/10.1021/ja00225a054
Evans RE (1964) 460. hydropyrimidines. part iii. reduction of amino-pyrimidines. J Chem Soc. https://doi.org/10.1039/JR9640002450
Mobley DL, Wymer KL, Lim NM, Guthrie JP (2014) Blind prediction of solvation free energies from the SAMPL4 challenge. J Comput Aided Mol Des 28(3):135. https://doi.org/10.1007/s10822-014-9718-2
Acknowledgements
DLM and CCB appreciate the financial support from the National Science Foundation (CHE 1352608) and the National Institutes of Health (1R01GM108889-01). CCB was supported financially by OpenEye Scientific Software to build this model during Summer 2017 and is now supported by a fellowship from The Molecular Sciences Software Institute under NSF Grant ACI-1547580. We are thankful for valuable conversations with OpenEye employees, the SAMPL6 organizers, and all challenge participants, and especially to Merck for its contributions to the experimental work in this challenge. AGS would like to thank Paul Hawkins, Christopher Bayly and Robert Tolbert as well as Anthony Nicholls and Matthew Geballe for many insightful discussions of \(pK_a\) and machine learning.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bannan, C.C., Mobley, D.L. & Skillman, A.G. SAMPL6 challenge results from \(pK_a\) predictions based on a general Gaussian process model. J Comput Aided Mol Des 32, 1165–1177 (2018). https://doi.org/10.1007/s10822-018-0169-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0169-z