Abstract
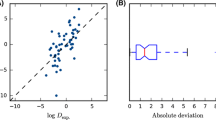
Here, we give an overview of the small molecule hydration portion of the SAMPL4 challenge, which focused on predicting hydration free energies for a series of 47 small molecules. These gas-to-water transfer free energies have in the past proven a valuable test of a variety of computational methods and force fields. Here, in contrast to some previous SAMPL challenges, we find a relatively wide range of methods perform quite well on this test set, with RMS errors in the 1.2 kcal/mol range for several of the best performing methods. Top-performers included a quantum mechanical approach with continuum solvent models and functional group corrections, alchemical molecular dynamics simulations with a classical all-atom force field, and a single-conformation Poisson–Boltzmann approach. While 1.2 kcal/mol is still a significant error, experimental hydration free energies covered a range of nearly 20 kcal/mol, so methods typically showed substantial predictive power. Here, a substantial new focus was on evaluation of error estimates, as predicting when a computational prediction is reliable versus unreliable has considerable practical value. We found, however, that in many cases errors are substantially underestimated, and that typically little effort has been invested in estimating likely error. We believe this is an important area for further research.






Similar content being viewed by others
Notes
For experimental measurements, the enantiomer composition could in principle be important if the hydration free energy is determined in part by a solubility measurement, because the solubility of a mixture of two enantiomers of a particular solute might be different than the solubility of either enantiomer alone. This mainly applies to racemic solids which form crystals with a racemic unit cell.
Some participants raised concerns about the experimental data for mannitol, which was well predicted by some submissions but poorly predicted by others, and was by far the most polar compound in the set. Because of these concerns, we also re-computed statistics for all methods without mannitol, to see how much this would affect conclusions. However, we found that this did not dramatically change the rank-ordering of methods by most metrics, at least not more than would be expected given the (substantial) bootstrapped error bars. Thus, given the lack of any definitive evidence to the contrary, and the fact that mannitol was not one of the least well-predicted compounds, we kept mannitol in the set.
References
Geballe MT, Guthrie JP (2012) The SAMPL3 blind prediction challenge: transfer energy overview. J Comput Aided Mol Des 26(5):489–496
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279
Klimovich P, Mobley DL (2010) Predicting hydration free energies using all-atom molecular dynamics simulations and multiple starting conformations. J Comput Aided Mol Des 24(4):307–316
Mobley DL, Bayly CI, Cooper MD, Dill KA, Dill KA (2009) Predictions of hydration free energies from all-atom molecular dynamics simulations. J Phys Chem B 113:4533–4537
Mobley DL, Liu S, Cerutti DS, Swope WC, Rice JE (2012) Alchemical prediction of hydration free energies for SAMPL. J Comput Aided Mol Des 26(5):551–562
Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779
Guthrie JP (2014) SAMPL4, a blind challenge for computational solvation free energies: the compounds considered. J Comput Aided Mol Des. doi:10.1007/s10822-014-9738-y
OpenEye Python Toolkits (2013)
Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA (2009) Small molecule hydration free energies in explicit solvent: an extensive test of fixed-charge atomistic simulations. J Chem Theory Comput 5(2):350–358
Mobley DL, Dumont É, Chodera JD, Dill K (2007) Comparison of charge models for fixed-charge force fields: small-molecule hydration free energies in explicit solvent. J Phys Chem B 111(9):2242–2254
Chodera JD, Noé F (2010) Probability distributions of molecular observables computed from Markov models. II. Uncertainties in observables and their time-evolution. J Chem Phys 133(10):105,102
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (1999) Numerical recipes in C, 2nd edn. Cambridge University Press, Cambridge
Yang W (2013) Personal Communication
Sandberg L (2013) Predicting hydration free energies with chemical accuracy: The SAMPL4 challenge. J Comput Aided Mol Des. doi:10.1007/s10822-014-9725-3
Ellingson BA, Geballe MT, Wlodek S, Bayly CI, Skillman AG, Nicholls A (2014) Efficient calculation of SAMPL4 hydration free energies using OMEGA, SZYBKI, QUACPACK, and Zap TK. J Comput Aided Mol Des. doi:10.1007/s10822-014-9720-8
Nicholls A, Wlodek S, Grant JA (2010) SAMPL2 and continuum modeling. J Comput Aided Mol Des 24(4):293–306
Jakalian A, Jack D, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM 1(BCC model): II. Parameterization and validation. J Comput Chem 23(16):1623–1641
Wang J, Wolf R, Caldwell J, Kollman P, Case D (2011) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174
Fennell CJ, Wymer KL, Mobley DL (2014) Polarized alcohol in condensed-phase and its role in small molecule hydration
Muddana HS, Sapra NV, Fenley AT, Gilson MK (2014) The SAMPL4 hydration challenge: evaluation of partial charge sets with explicit-water molecular dynamics simulations. J Comput Aided Mol Des. doi:10.1007/s10822-014-9714-6
Canzar S, El-Kebir M, Pool R, Elbassioni K, Malde AK, Mark AE, Geerke DP, Stougie L, Klau GW (2013) Charge group partitioning in biomolecular simulation. J Comput Biol 20(3):188–198
Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE (2011) An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theory Comput 7(12):4026–4037
Hawkins GD, Giesen DJ, Lynch GC, Chambers CC, Rossi I, Storer JW, Li J, Zhu T, Thompson J, Winget P, Lynch BJ AMSOL. http://comp.chem.umn.edu/amsol/
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52(7):1757–1768
Hawkins PCD, Nicholls A (2012) Conformer generation with OMEGA: learning from the data set and the analysis of failures. J Chem Inf Model 52(11):2919–2936
Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge structural database. J Chem Inf Model 50(4):572–584
Hogues H, Sulea T, Purisima EO (2014) Exhaustive docking and solvated interaction energy scoring: lessons learned from the SAMPL4 challenge. J Comput Aided Mol Des. doi:10.1007/s10822-014-9715-5
Klamt A, Eckert F, Diedenhofen M (2009) Prediction of the free energy of hydration of a challenging set of pesticide-like compounds. J Phys Chem B 113(14):4508–4510
Reinisch J, Klamt A (2014) Prediction of free energies of hydration with COSMO-RS on the SAMPL4 data set. J Comput Aided Mol Des. doi:10.1007/s10822-013-9701-3
Sulea T, Purisima EO (2011) Predicting hydration free energies of polychlorinated aromatic compounds from the SAMPL-3 data set with FiSH and LIE models. J Comput Aided Mol Des 26(5):661–667
Li L, Dill KA, Fennell CJ (2014) Hydration assembly tests in the SAMPL4 challenge. J Comput Aided Mol Des. doi:10.1007/s10822-014-9712-8
Acknowledgments
We acknowledge the financial support of the National Institutes of Health (1R15GM096257-01A1), and computing support from the UCI GreenPlanet cluster, supported in part by NSF Grant CHE-0840513. We also thank J. Peter Guthrie for help with sorting out structure and naming confusion in SAMPL preparation, several SAMPL participants including Jens Reinisch and Samuel Genheden for helpful exchanges on issues with the guaiacol series, and Andreas Klamt for help on data relating to 1-benzylimidazole. We also thank OpenEye for their support of the SAMPL meeting and for running the web server, and Matt Geballe (OpenEye) for help managing the web site and automated submission system.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Mobley, D.L., Wymer, K.L., Lim, N.M. et al. Blind prediction of solvation free energies from the SAMPL4 challenge. J Comput Aided Mol Des 28, 135–150 (2014). https://doi.org/10.1007/s10822-014-9718-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9718-2