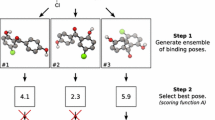
Abstract
Several combinations of docking software and scoring functions were evaluated for their ability to predict the binding of a dataset of potential HIV integrase inhibitors. We found that different docking software were appropriate for each one of the three binding sites considered (LEDGF, Y3 and fragment sites), and the most suitable two docking protocols, involving Glide SP and Gold ChemScore, were selected using a training set of compounds identified from the structural data available. These protocols could successfully predict respectively 20.0 and 23.6 % of the HIV integrase binders, all of them being present in the LEDGF site. When a different analysis of the results was carried out by removing all alternate isomers of binders from the set, our predictions were dramatically improved, with an overall ROC AUC of 0.73 and enrichment factor at 10 % of 2.89 for the prediction obtained using Gold ChemScore. This study highlighted the ability of the selected docking protocols to correctly position in most cases the ortho-alkoxy-carboxylate core functional group of the ligands in the corresponding binding site, but also their difficulties to correctly rank the docking poses.







Similar content being viewed by others
References
Skillman AG (2012) SAMPL3: blinded prediction of host-guest binding affinities, hydration free energies, and trypsin inhibitors. J Comput Aided Mol Des 26(5):473–474. doi:10.1007/s10822-012-9580-z
Newman J, Dolezal O, Fazio V, Caradoc-Davies T, Peat TS (2012) The DINGO dataset: a comprehensive set of data for the SAMPL challenge. J Comput Aided Mol Des 26(5):497–503. doi:10.1007/s10822-011-9521-2
Mobley DL, Liu S, Lim NM, Wymer KL, Perryman AL, Forli S, Deng N, Su J, Branson K, Olson AJ (2014) Blind prediction of HIV integrase binding from the SAMPL4 challenge. J Comput Aided Mol Des
Peat TS, Dolezal O, Newman J, Mobley D, Deadman JJ (2014) Interrogating HIV integrase for compounds that bind—a SAMPL challenge
Mtifiot M, Marchand C, Pommier Y (2013) HIV integrase inhibitors: 20-year landmark and challenges. Adv Pharmacol 67:75–105. doi:10.1016/B978-0-12-405880-4.00003-2
Hazuda DJ (2012) HIV integrase as a target for antiretroviral therapy. Curr Opin HIV AIDS 7(5):383–389. doi:10.1097/COH.0b013e3283567309
Quashie PK, Sloan RD, Wainberg MA (2012) Novel therapeutic strategies targeting HIV integrase. BMC Med 10:34. doi:10.1186/1741-7015-10-34
Surpateanu G, Iorga BI (2012) Evaluation of docking performance in a blinded virtual screening of fragment-like trypsin inhibitors. J Comput Aided Mol Des 26(5):595–601. doi:10.1007/s10822-011-9526-x
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res Suppl 28(1):235–242
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10(12):980. doi:10.1038/nsb1203-980
Rhodes DI, Peat TS, Vandegraaff N, Jeevarajah D, Le G, Jones ED, Smith JA, Coates JAV, Winfield LJ, Thienthong N, Newman J, Lucent D, Ryan JH, Savage GP, Francis CL, Deadman JJ (2011) Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design. Antivir Chem Chemother 21(4):155–168. doi:10.3851/IMP1716
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. doi:10.1002/jcc.21256
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein-ligand docking using GOLD. Proteins 52(4):609–623. doi:10.1002/prot.10465
Acknowledgments
Our laboratory is a member of the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (ANR-10-LABX-33). We would like to thank the SAMPL4 organizers, with a special mention to David L. Mobley, for providing the experimental data required for the evaluation of our predictions, as well as for the alternate analysis of the virtual screening results. The pertinent comments and suggestions of the manuscript reviewers are also kindly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Colas, C., Iorga, B.I. Virtual screening of the SAMPL4 blinded HIV integrase inhibitors dataset. J Comput Aided Mol Des 28, 455–462 (2014). https://doi.org/10.1007/s10822-014-9707-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9707-5