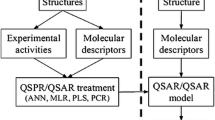
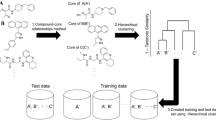
Abstract
This article is about the hierarchical quantitative structure–activity relationship technology (HiT QSAR) based on the Simplex representation of molecular structure (SiRMS) and its application for different QSAR/QSP(property)R tasks. The essence of this technology is a sequential solution (with the use of the information obtained on the previous steps) to the QSAR problem by the series of enhanced models of molecular structure description [from one dimensional (1D) to four dimensional (4D)]. It is a system of permanently improved solutions. In the SiRMS approach, every molecule is represented as a system of different simplexes (tetratomic fragments with fixed composition, structure, chirality and symmetry). The level of simplex descriptors detailing increases consecutively from the 1D to 4D representation of the molecular structure. The advantages of the approach reported here are the absence of “molecular alignment” problems, consideration of different physical–chemical properties of atoms (e.g. charge, lipophilicity, etc.), the high adequacy and good interpretability of obtained models and clear ways for molecular design. The efficiency of the HiT QSAR approach is demonstrated by comparing it with the most popular modern QSAR approaches on two representative examination sets. The examples of successful application of the HiT QSAR for various QSAR/QSPR investigations on the different levels (1D–4D) of the molecular structure description are also highlighted. The reliability of developed QSAR models as predictive virtual screening tools and their ability to serve as the base of directed drug design was validated by subsequent synthetic and biological experiments, among others. The HiT QSAR is realized as a complex of computer programs known as HiT QSAR software that also includes a powerful statistical block and a number of useful utilities.






Similar content being viewed by others
Notes
A preliminary step is that the list of structural parameters must be well-organized on a defined principle (for example, lexicographic).
This classification is offered by the authors of Cerius 2.
The authors express their sincere gratitude to Prof. J. Leszczynski, Dr. L. Gorb and Dr. M. Quasim for fruitful cooperation during the development of this task.
The authors express their sincere gratitude to Dr. M. Schmidtke, Prof. P. Wutzler, Dr. V. Makarov, Dr. O. Riabova, Mr. N. Kovdienko and Mr. A. Hromov for their most fruitful cooperation that made the development of this task possible.
The authors express their sincere gratitude to Dr. M. Schmidtke, Prof. P. Wutzler, Dr. V. Makarov, Dr. O. Riabova and Ms. Volineckaya for their fruitful cooperation that made possible the development of this task.
The authors express their sincere gratitude to Prof. G.L. Kamalov, Dr. S.A. Kotlyar and Dr. G.N. Chuprin for fruitful cooperation during the development of this task.
The authors express their sincere gratitude to Dr. V.P. Lozitsky, Dr. R.N. Lozytska and Dr. A.S. Fedtchouk for their fruitful cooperation during the development of this task.
The anti-influenza and antiherpetic investigations described were carried out as a result of fruitful cooperation with Dr. V.P. Lozitsky, Dr. R.N. Lozytska and Dr. A.S. Fedtchouk, Dr. T.L. Gridina, Dr. S. Basok, Dr. D. Chikhichin, Mr. V. Chelombitko and Dr. J.-J. Vanden Eynde. The authors express their sincere gratitude to all of these colleagues.
In this and anti-iherpetic research 1D modeling was not carried out.
We are aware that these models can approximate not only the variation in activity but also the variation in experimental errors. The high values of the R 2 test can be explained by the fact that test compounds are very similar to the training ones, that there are only few compounds in test set, by the high quality of the obtained models, by simple good luck and/or by a combination of all these factors.
Abbreviations
- A/I/EVS:
-
Automatic/interactive/evolutionary variables selection
- ACE:
-
Angiotensin converting enzyme
- AchE:
-
Acetylcholinesterase
- CoMFA:
-
Comparative molecular fields analysis QSAR approach
- CoMSIA:
-
Comparative molecular similarity indexes analysis QSAR approach
- DA:
-
Applicability domain
- DSTP:
-
Dispirotripiperazine
- EVA:
-
Eigenvalue analysis QSAR approach
- GA:
-
Genetic algorithm
- HiT QSAR:
-
Hierarchical QSAR technology
- HQSAR:
-
Hologram QSAR approach
- HRV:
-
Human rhinovirus
- HSV:
-
Herpes simplex virus
- MLR:
-
Multiple linear regression statistical method
- PLS:
-
Partial least squares or projection on latent structures statistical method
- Q 2 :
-
Cross-validation determination coefficient
- QSAR/QSPR:
-
Quantitative structure–activity/property relationship
- R 2 :
-
Determination coefficient for training set
- R 2 test :
-
Determination coefficient for test set
- SD:
-
Simplex descriptor
- SI:
-
Selectivity index
- SiRMS:
-
Simplex representation of the molecular structure QSAR approach
- TV:
-
Trend-vector statistical method
References
Ooms F (2000) Curr Med Chem 7(2):141
Thomas G (2008) Medicinal chemistry: an introduction, 2nd edn. Wiley
Artemenko AG, Muratov EN, Kuz’min VE, Kovdienko NA, Hromov AI, Makarov VA, Riabova OB, Wutzler P, Schmidtke M (2007) J Antimicrob Chemother 60(1):68
Bailey TR, Diana GD, Kowalczyk PJ, Akullian V, Eissenstat MA, Cutcliffe D, Mallamo JP, Carabateas PM, Pevear DC (1992) J Med Chem 35(24):4628
Butina D, Gola JMR (2004) J Chem Inf Comput Sci 43:837
de Jonge MR, Koymans LM, Vinkers HM, Daeyaert FF, Heeres J, Lewi PJ, Janssen PA (2005) J Med Chem 48(6):2176
Jenssen H, Gutteberg TJ, Lejon T (2005) J Pept Sci 11(2):97
Kovatcheva A, Golbraikh A, Oloff S, Xiao Y, Zheng W, Wolschann P, Buchbauer G, Tropsha A (2004) J Chem Inf Comput Sci 44:582
Kubinyi H (1990) J Cancer Res Clin Oncol 116:529
Kuz’min VE, Artemenko AG, Lozitska RN, Fedtchouk AS, Lozitsky VP, Muratov EN, Mescheriakov AK (2005) SAR QSAR Environ Res 16(3):219
Kuz’min VE, Artemenko AG, Muratov EN, Volineckaya IL, Makarov VA, Riabova OB, Wutzler P, Schmidtke M (2007) J Med Chem 50:4205
Muratov EN, Artemenko AG, Kuz’min VE, Lozitsky VP, Fedchuk AS, Lozitska RN, Boschenko YA, Gridina TL (2005) Antiviral Res 65(3):A62
Verma RP, Hansch C (2006) Curr Med Chem 13(4):423
Zhang S, Golbraikh A, Tropsha A (2006) J Med Chem 49:2713
Selassie CD (2003) In: Abraham DJ (ed) Burger’s medicinal chemistry and drug discovery, 6th edn, vol 1. Wiley, New York
Cramer RD, Patterson DI, Bunce JD (1988) J Am Chem Soc 110:5959
Doweyko AM (1988) J Math Chem 31:1396
Klebe G, Abraham U, Mietzner T (1994) J Med Chem 37:4130
Kuz’min VE, Artemenko AG, Kovdienko NA, Tetko IV, Livingstone DJ (2000) J Mol Model 6:517
Seel M, Turner DB, Wilett P (1999) QSAR 18:245
Pavan M, Consonni V, Gramatica P, Todeschini R (2006) In: Partial order in environmental sciences and chemistry. Springer, Berlin, pp 181–217
Baurin N, Mozziconacci JC, Arnoult E, Chavatte P, Marot C, Morin-Allory L (2004) J Chem Inf Model 44(1):276
Vedani A, Dobler M (2000) Prog Drug Res 55:107
Kuz’min VE, Artemenko AG, Polischuk PG, Muratov EN, Hromov AI, Liahovskiy AV, Andronati SA, Makan SY (2005) J Mol Model 11(6):457
Kuz’min VE, Artemenko AG, Muratov EN, Lozitsky VP, Fedchuk AS, Lozitska RN, Boschenko YA, Gridina TL (2005) Antiviral Res 65(3):A70
Artemenko A, Kuz’min V, Muratov E, Fedchuk A, Lozitsky V, Gridina T, Lozytska R, Basok S, Chikhichin D (2007) Antiviral Res 74:A76
Artemenko A, Muratov E, Kuz’min V, Fedtchuk A, Mykhaylovska N, Lesyk R, Zimenkovsky B (2006) Clin Microbiol Infect 12(4):1557
Artemenko A, Muratov E, Kuz’min V, Koroleva L, Silnikov V, Lozitsky V, Fedchuk A (2006) Antiviral Res 70:A43
Artemenko AG, Kuz’min VE, Muratov EN, Lozitsky VP, Fedchuk AS, Lozitska RN, Boschenko YA, Gridina TL (2005) Antiviral Res 65(3):A77
Kuz’min VE, Artemenko AG, Lozitsky VP, Muratov EN, Fedtchouk AS, Dyachenko NS, Nosach LN, Gridina TL, Shitikova LI, Mudrik LM, Mescheriakov AK, Chelombitko VA, Zheltvay AI, Vanden Eynde J-J (2002) Acta Biochim Pol 49:157
Kuz’min VE, Artemenko AG, Muratov EN, Volineckaya IL, Makarov VA, Riabova OB, Wutzler P, Schmidtke M (2007) Antiviral Res 74:A49
Muratov E, Artemenko A, Kuz’min V, Konup I, Konup L, Kotlyar S, Kamalov G, Fedtchuk A, Mykhaylovska N (2006) Clin Microbiol Infect 12(4):1558
Muratov EN, Kuz’min VE, Artemenko AG, Makarov VA, Riabova OB, Wutzler P, Schmidtke M, Lozitsky V, Fedchuk A (2006) Antiviral Res 70:A77
QSAR; Expert; Group (2004) The report from the expert group on (quantitative) structure–activity relationships [(Q)SARs] on the principles for the validation of (Q)SARs.; 49; Organisation for Economic Co-operation and Development, Paris
Kuz’min VE (1995) Zh Strucur Khim 36:873
Jolly WL, Perry WB (1973) J Am Chem Soc 95:5442
Wang R, Fu Y, Lai L (1997) J Chem Inf Comput Sci 37:615
Ioffe BV (1983) Chemistry refractometric methods, 3rd edn. Himiya, Leningrad
Burkert U, Allinger N (1982) Molecular mechanics. ACS Publication, Washington D.C.
Marple SL Jr (1987) Digital spectral analysis with applications. Prentice-Hall, New York
Östergard PRJ (2002) Discrete Appl Math 120:195
Muratov EN (2004) Quantitative evaluation of the structural factors influence on the properties of nitrogen-, oxygen- and sulfur-containing macroheterocycles. A.V. Bogatsky Physical-Chemical Institute, Odessa
Bodor N, Buchwald P (2000) Med Res Rev 20(1):58
Cronin MTD, Schultz TW (2003) J Mol Struct (Theochem) 622:39
Kubinyi H (1996) J Chemometr 10:119
Lindgren F, Geladi P, Rannar S, Wold S (1994) J Chemometr 8:349
Rannar S, Lindgren F, Geladi P, Wold S (1994) J Chemometr 8:111
Rogers D, Hopfinger AJ (1994) J Chem Inf Comput Sci 34(4):854
Carhart RE, Smith DH, Venkataraghavan R (1985) J Chem Inf Comput Sci 25:64
Vitiuk NV, Kuz’min VE (1994) Zh Anal Khim 49:165
Wold S, Antti H, Lindgren F, Ohman J (1998) Chemometr Intell Lab Syst 44:175
Trygg J, Wold S (2002) J Chemometr 16:119
Neter J, Kutner MH, Wasseman W, Nachtsheim C (1996) Applied linear statistical models. McGraw-Hill, New York
Meloun M, Militku J, Hill M, Brereton MG (2002) Analyst 127:433
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) Altern Lab Anim 33:445
Sutherland JJ, O’Brien LA, Weaver DF (2004) J Med Chem 47:5541
Heritage TV, Ferguson AM, Turner DB, Willett P (1998) Perspect Drug Discovery Des 11:381
Free SM, Wilson JW (1964) J Med Chem 7:395
Kuz’min VE, Lozitsky VP, Kamalov GL, Lozitskaya RN, Zheltvay AI, Fedtchouk AS, Kryzhanovsky DN (2000) Acta Biochim Pol 47:867
Acknowledgments
This work was partially supported by a grant of the President of Ukraine (President of Ukraine grant for young investigators GP/F11/0115), the Science & Technology Center in Ukraine (STCU project 3147) and INTAS foundation (INTAS Grant 97-31528).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kuz’min, V.E., Artemenko, A.G. & Muratov, E.N. Hierarchical QSAR technology based on the Simplex representation of molecular structure. J Comput Aided Mol Des 22, 403–421 (2008). https://doi.org/10.1007/s10822-008-9179-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-008-9179-6