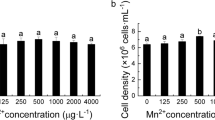
Abstract
Nitzschia ZJU1, which originated from the diatom Nitzschia sp. (ash free dry weight of biomass was 0.12 g L−1 and its lipid content was 13.34 %) after 60Co-γ ray irradiation and domestication at high-salinity, was re-mutated by 137Cs-γ irradiation to increase lipid productivity. The lipid yields of the new mutants Nitzschia ZJU2 and Nitzschia ZJU3, which were selected by Nile Red fluorescence, were increased from 209.9 mg L−1 (for the original Nitzschia ZJU1) to 245.5 mg L−1 and 311.6 mg L−1, respectively. The lipid content of the two strains increased to 64.42 and 62.61 % of ash free dry weight, respectively, when the cells were cultured with nitrogen and silicon deprivation. It was found that 3,063 reads of genetic expression including Acetyl-CoA carboxylase and other important genes in lipid synthesis pathway were up-regulated and 4,598 reads of genetic expression were down-regulated in mutant Nitzschia ZJU2, when the cells were cultured in optimized growth medium without nitrogen and silicon. The gene expression levels of ATP-binding cassette transporters, arginine and proline metabolism, and proteasome in metabolic pathways were up-regulated to different degrees in mutant Nitzschia ZJU2 under the same cultivation condition.







Similar content being viewed by others
References
Baud S, Wuillème S, Dubreucq B, De Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52:405–419
Bender SJ, Durkin CA, Berthiaume CT, Morales RL, Armbrust EV (2014) Transcriptional responses of three model diatoms to nitrate limitation of growth. Front Mar Sci 1:1–15
Bligh E, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Borowitzka LJ, Volcani B (1977) Role of silicon in diatom metabolism. Arch Microbiol 112:147–152
Cheng J, Huang Y, Feng J, Sun J, Zhou J, Cen K (2013a) Improving CO2 fixation efficiency by optimizing Chlorella PY-ZU1 culture conditions in sequential bioreactors. Bioresource Technol 144:321–327
Cheng J, Huang Y, Feng J, Sun J, Zhou J, Cen K (2013b) Mutate Chlorella sp. by nuclear irradiation to fix high concentrations of CO2. Bioresource Technol 136:496–501
Cheng J, Feng J, Sun J, Huang Y, Zhou J, Cen K (2014a) Enhancing the lipid content of the diatom Nitzschia sp. by 60Co-γ irradiation mutation and high-salinity domestication. Energy (Under review)
Cheng R-L, Feng J, Zhang B-X, Huang Y, Cheng J, Zhang C-X (2014b) Transcriptome and gene expression analysis of an oleaginous diatom under different salinity conditions. Bioenerg Res 7:192–205
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25:294–306
Cooksey KE, Guckert JB, Williams SA, Callis PR (1987) Fluorometric determination of the neutral lipid content of microalgal cells using Nile Red. J Microbiol Meth 6:333–345
Courchesne NEMM, Parisien A, Wang B, Lan CQ (2009) Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol 141:31–41
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S (2000) Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A 97:6487–6492
de Castro AS, Garcia VMT (2005) Growth and biochemical composition of the diatom Chaetoceros cf. wighamii Brightwell under different temperature, salinity and carbon dioxide levels. I. Protein, carbohydrates and lipids. Aquaculture 246:405–412
Elsey D, Jameson D, Raleigh B, Cooney MJ (2007) Fluorescent measurement of microalgal neutral lipids. J Microbiol Meth 68:639–642
Garnham GW, Codd GA, Gadd GM (1993) Uptake of cobalt and cesium by microalgal-and cyanobacterial-clay mixtures. Microbial Ecol 25:71–82
Ge F, Huang W, Chen Z, Zhang C, Xiong Q, Bowler C, Yang J, Xu J, Hu H (2014) Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell 26:1681–1697
Hildebrand M, Dahlin K (2000) Nitrate transporter genes from the diatom Cylindrotheca fusiformis (Bacillariophyceae): mRNA leves controlled by nitrogen source and by the cell cycle. J Phycol 36:702–713
Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54:621–639
Illman AM, Scragg AH, Shales SW (2000) Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme Microb Tech 27:631–635
Khozin-Goldberg I, Cohen Z (2011) Unraveling algal lipid metabolism: Recent advances in gene identification. Biochimie 93:91–100
Kovacs E, Keresztes A (2002) Effect of gamma and UV-B/C radiation on plant cells. Micron 33:199–210
Kranz RG, Gabbert KK, Madigan MT (1997) Positive selection systems for discovery of novel polyester biosynthesis genes based on fatty acid detoxification. Appl Environ Microb 63:3010–3013
Liu Z-Y, Wang G-C, Zhou B-C (2008) Effect of iron on growth and lipid accumulation in Chlorella vulgaris. Bioresource Technol 99:4717–4722
Lynn SG, Kilham SS, Kreeger DA, Interlandi SJ (2000) Effect of nutrient availability on the biochemical and elemental stoichiometry in the freshwater diatom Stephanodiscus minutulus (Bacillariophyceae). J Phycol 36:510–522
Mishra NP, Mishra RK, Singhal GS (1993) Changes in the activities of anti-oxidant enzymes during exposure of intact wheat leaves to strong visible light at different temperatures in the presence of protein synthesis inhibitors. Plant Physiol 102:903–910
Mock T, Samanta MP, Iverson V, Berthiaume C, Robison M, Holtermann K, Durkin C, BonDurant SS, Richmond K, Rodesch M et al (2008) Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc Nat Acad Sci 105:1579–1584
Moheimani N, Borowitzka M, Isdepsky A, Sing S (2013) Standard methods for measuring growth of algae and their composition. In: Moheimani NR, Borowitzka MA (eds) Algae for Biofuels and Energy. Springer, Dordrecht, pp 265–284
Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LGG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38:D822–D827
Schwender JOR, Ohlrogge JB (2002) Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol 130:347–361
Walker D (2009) Biofuels, facts, fantasy, and feasibility. J Appl Phycol 21:509–517
Wang ZP, Zhao Y (2005) Morphological reversion of Spirulina (Arthrospira) platensis (Cyanophyta): from linear to helical. J Phycol 41:622–628
Weldy CS, Huesemann M (2007) Lipid production by Dunaliella salina in batch culture: effects of nitrogen limitation and light intensity. US DOE J Undergraduate Res 7:115–122
Widjaja A, Chien C-C, Ju Y-H (2009) Study of increasing lipid production from fresh water microalgae Chlorella vulgaris. J Taiwan Inst Chem E 40:13–20
Zhu CJ, Lee YK (1997) Determination of biomass dry weight of marine microalgae. J Appl Phycol 9:189–194
Acknowledgement
This work was supported by the National Natural Science Foundation of China (51176163), National High Technology R&D Program of China (2012AA050101), International Sci. & Tech. Cooperation Program of China (2012DFG61770), National Key Technology R&D Program of China (2011BAD14B02), Zhejiang Provincial Natural Science Foundation of China (LR14E060002), Program for New Century Excellent Talents in University (NCET-11-0446), Specialized Research Fund for the Doctoral Program of Higher Education (20110101110021), Science and Technology Project of Guangxi Province (1346011–1).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1
Compositions of growth medium F for diatom Nitzschia ZJU1 (XLSX 10 kb)
Supplementary Table S2
Genes differentially expressed in Nitzschia ZJU2 under silicon starvation. (XLSX 286 kb)
Supplementary Table S3
Genes differentially expressed in Nitzschia ZJU2 under nitrogen starvation (XLSX 410 kb)
Supplementary Table S4
Differentially expressed genes in Nitzschia ZJU2 under nitrogen and silicon starvation. (XLSX 631 kb)
Supplementary Table S5
Differentially expressed genes related to C and N metabolism in Nitzschia ZJU2. (XLSX 21 kb)
Rights and permissions
About this article
Cite this article
Feng, J., Cheng, J., Cheng, R. et al. Screening the diatom Nitzschia sp. re-mutated by 137Cs-γ irradiation and optimizing growth conditions to increase lipid productivity. J Appl Phycol 27, 661–672 (2015). https://doi.org/10.1007/s10811-014-0367-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-014-0367-6