Abstract
Diversity of Cyanophyceae/cyanobacteria is expressed by their morphological, biochemical and physiological properties, which enable them to settle and persist in a wide range of habitats. Their diverse morphology determined their taxonomic distinction based on phenotypic properties. The oxygenic photosynthesis which characterizes cyanobacteria and their sharing of ecological niches with eukaryotic algae, prompted their treatment in the phycological circles, where they were called blue-green algae, although their prokaryotic nature, akin to bacteria, has been recognized for over a century. The cyanobacteria are named under Botanical and Bacteriological Codes, and the usage of both systems at the same time causes considerable confusion as the rules of the Botanical Code are quite different from those of the Bacteriological one. Herbarium collections are perfect subjects for intensive phylogenetic studies and therefore can contribute to discussions on the traditional and newly emerging concepts of species and speciation in prokaryotes. This article reviews the present status of the taxonomy of cyanobacteria, describes earlier, classical and recent taxonomic approaches and the trends for future, emphasizing improvements in methodology as major catalysts for the progress of this field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The history of cyanobacterial systematics has been unusually tumultuous. The metabolic property of oxygenic photosynthesis which characterizes cyanobacteria and their sharing of ecological niches with eukaryotic algae, prompted their treatment in the phycological circles, where they were called blue-green algae, although their prokaryotic nature, akin to bacteria, has been recognized for over a century. The main problem in cyanobacterial systematics is that the sexual reproduction in cyanobacteria is not known to date. It results in a number of consequences: (i) traditional species concepts (Mayr, 1982) cannot be applied; (ii) strains evolve and undergo evolution; (iii) the number of transitional forms and ecotypes is numerous (Moore et al., 1998; Rippka et al., 2000); (iv) natural populations differ from cultured strains which change drastically their morphology under cultural conditions (Palinska et al., 1996). In spite of it, many important morphologically and ecophysiologically stable units exist in nature, and they occur repeatedly in distant localities under similar ecological situations. They are important for ecologists, and thus, the traditional morphological concept of species is still commonly used in ecological studies (Palinska & Surosz, 2008).
Blue-green algae in botanical tradition have been distinguished on the basis of phenotypic properties. Thuret (1875), Bornet & Flahaut (1887, 1888a, b) and Gomont (1892) wrote the first comprehensive taxonomic monographs for blue-greens, recognized by phycologists as a later starting point in taxonomic referencing. The basic unit of that system is a species as defined in botany and zoology (e.g. Mayr, 1982). Geitler (1932) provided an updated taxonomic review and determination manual that recognized 1,300 species, classified into 145 genera, 20 families and 3 orders. Geitler’s work relied on morphology of field-collected specimens and his classification system marks the beginning of the modern era of cyanobacterial systematics, recognized both by phycologists and microbiologists. It has formed the basis of numerous revised systems proposed since then, including those of Elenkin (1938, 1949), Desikachary (1959), Fritsch (1959), Starmach (1966), Kondrateva (1968), Bourelly (1970) and Golubic (1976). These systems share the view that the systematics of cyanophytes should be based on traditional botanical criteria, a view sometimes referred to as the “Geitlerian” approach.
In the period 1956–1981, an alternative system was developed by Drouet and Daily (summarized in Drouet, 1981), that drastically reduced the number of genera and species of blue-green algae. It was based on the hypothesis that the many morphological differences seen in natural samples of cyanophytes are ephemeral and that numerous “species” of cyanobacteria are actually different “ecophenes” of true taxa. However, it was shown that this system does not reflect the true genetic diversity among blue-greens and was never fully accepted.
Waterbury & Stanier (1977), Krumbein (1979) and Rippka et al. (1979) proposed that the systematic treatment of cyanobacteria should be based on bacteriological criteria on the ground that they are unquestionably prokaryotes. The basic taxonomic unit in bacteriological treatment is an axenic-cultured strain, whereas the species becomes a conceptual construct based on comparison of a number of similar strains. In revising the cyanobacterial genera, Stanier school relied largely on morphological properties and used Geitlerian designations, but altered many generic definitions in accordance with properties expressed in culture (Castenholz & Waterbury, 1989). In using this approach, many bacteriologists avoid phenotypic species description. Accordingly, cultured cyanobacteria are usually assigned the name of genus with a strain code. Moreover, bacteriological approach leads to gross underestimation of the cyanobacteria diversity in nature. The current edition of the Bergey’s Manual of Systematic Bacteriology includes compiled information from both bacteriological and phycological sources (Castenholz, 2001).
Another update and revision of cyanobacterial system, which includes ultrastructural properties. is currently underway (Anagnostidis & Komarek, 1985; Komarek & Anagnostidis, 1999, 2005; Komárek, 2013). Komárek & Anagnostidis (1999, 2005) have developed a formal system being a compromise between Bacteriological and Botanical approaches. Its nomenclature is based on botanical taxonomic criteria, but it also utilizes bacteriological and molecular information.
This article reviews the present status of the taxonomy of cyanobacteria, describes historical and more recent taxonomic approaches and presents the trends for future, highlighting improvements and developments in methodology as major promoters for the progress of this scientific discipline.
Cyanophytes/cyanobacteria under the botanical and the bacteriological codes: a comparison
Classical taxonomy represents a body of work that has accumulated over past 250 years, since the introduction of the binominal naming system by Linnaeus in the 1750s. A crucial component of current practice in taxonomy of cyanobacteria as well as in general taxonomy is the concept of the type specimen that serves as the central reference for comparisons. Designating a type specimen is required when a new species is named, and these are usually deposited in collections and herbaria. However, the system depends heavily on specialists whose knowledge is frequently lost when they are not active any more.
Today, there are still different “species” concepts and definitions in different groups of organisms. There are different Codes of Nomenclature guiding the relationships among taxa. Neither of these rules requires documentation of phylogenetic interrelationships among taxa, although such background was always desirable.
The “Cyanophytes” were traditionally classified as “blue-green algae”, with respect to their morphological diversity and size corresponding to other microalgae, as well as to their function in natural biotopes. Therefore, they were for a long period in the field of interest of botanists (phycologists) and ecologists, and treated as microscopic plant organisms. The detailed cytological and biochemical studies performed by bacteriologists on axenic culture strains led to the proposal to change the common name of “Cyanophyceae” into “cyanobacteria”, and to the opinion, that they should be classified as bacteria and nomenclatorially ruled by the International Code of Nomenclature of Bacteria (Stanier et al., 1978). The introduction of important model strains into the laboratory practice supported this approach (Castenholz, 2001).
The coexistence of two independent Codes Botanical (since 2012: the International Code of Nomenclature for algae, fungi and plants; Oren, 2014) and Bacteriological existing for one and the same group of organisms causes immense problems (Oren, 2014; Oren & Garrity, 2014). Names of cyanobacteria described and validly published as blue-green algae under the International Code of Botanical Nomenclature have no standing in bacterial nomenclature, unless they are again described under the Rules of the Bacteriological Code.
The rules of the Botanical Code are quite different from those of the Bacteriological Code, and this makes reconciliation between the botanical and the bacteriological nomenclature systems quite problematic. However, such reconciliation is urgently needed (Oren, 2004, 2011). The phycological practice concerned with populations in nature relies on the Botanical Code, using preserved-type specimens as taxonomic reference and the rule of priority in naming. In contrast, microbiologists use axenic cultures as basic taxonomic unit and a continuously revised approval of valid names, relying on the Bacteriological Code. Furthermore, according to the botanical approach, species names can be validly published in any journal, and the existing botanical nomenclatural information is widely scattered. To establish whether a species is novel to science, the literature to be checked extends over more than a hundred years. In contrast, IJSEM/IJSB is the only platform for bacteriological nomenclature. From the situation that two Codes guide the taxonomy of cyanobacteria and from the endeavour to come to any compromise of the bacteriological and botanical approaches, several proposals leading to the compatible nomenclatural procedures using the both Bacteriological and Botanical Codes were published. The important steps in this effort are the compromise proposals of Friedmann & Borowitzka (1982) and, recently, the corresponding chapters in the both editions of Bergey’s Manual of Systematic Bacteriology (Castenholz & Waterbury, 1989; Castenholz, 2001), from which the majority of proposed principles should be accepted in the nomenclature treatment of Cyanophyta/cyanobacteria. However, two Codes of Nomenclature (ICNB and ICBN), which are applicable for oxyphototrophic prokaryotes, none without obstacles, exist to date. That is why it is extremely important to establish minimal standards for the description of new cyanobacterial species and genera which will be acceptable to the botanical and bacteriological authorities at the same time. This should be followed by the publication of an “Approved List of Names of Cyanobacteria” in IJSEM. The ultimate goal is to achieve a consensus nomenclature that is acceptable both to bacteriologists and to botanists, anticipating the future implementation of a universal “Biocode” that would regulate the nomenclature of all organisms living on Earth (Oren, 2004).
The problem of species usage in cyanobacteria
A principal aim of systematics is to discover, describe and classify the diversity of living organisms. Systematists have concluded that the basic unit of biological diversity is the species. However, there is no widely accepted concept of species for prokaryotes, and assignment of isolates to species is based on measures of phenotypic or genome similarity. The current methods for defining prokaryotic species are inadequate and incapable of keeping pace with the levels of diversity that is being uncovered in nature (Stackebrandt et al., 2002).
Bacterial systematics has not yet reached a consensus for defining the fundamental unit of biological diversity, the species. The past half-century of bacterial systematics has been characterized by improvements in methods for demarcating species as phenotypic and genetic clusters, but species demarcation has not been guided by a theory-based concept of species. There is a discrepancy between operational (practical) species definition and theoretical species concept (Cohan, 2004). Species definition tends to be more arbitrary and focuses in practical necessity (Stackebrandt et al., 2002).
Prokaryotic species are currently characterized using a polyphasic approach that incorporates genotypic and phenotypic properties (Vandamme et al., 1996; Stackebrandt et al., 2002). Since the 1970s, the basis of genotypic characterization has been the measurement of overall genetic similarity among isolates, assessed by the degree to which their genomes hybridize under standard conditions [DNA–DNA hybridization (DDH)]. Here, both similarities in gene content and nucleotide similarity of shared genes contribute to a measure of the overall relatedness of their genomes. The recommendation to delineate species using a 70% DNA–DNA binding criterion does not correspond to a theory-based concept of what properties a species should have, but was calibrated empirically to yield many of the phenotype-based species already recognized at the time of its setting up. Using ribosomal RNA gene sequence similarity, pioneered by Woese & Fox (1977), systematists have invented robust method of creating evolutionary trees. With the help of this common method, bacteriologists widely recognize that bacterial diversity is organized into discrete phenotypic and genetic clusters, which are separated by phenotypic and genetic gaps, and these clusters are recognized as species (Dawson & Sneath, 1985).
Although the advantages of the direct genotypic approaches are clear, classification by rRNA gene sequence alone—an increasingly common practice—is unsatisfying for several reasons. The rRNA gene sequences often lack resolution when compared with DDH. Whereas isolates that have less than 97% rRNA gene sequence similarity usually share less than 70% DDH and belong to different species, isolates that have more than 97% identity might or might not meet the 70% DDH criterion for inclusion in the same species (Fox et al., 1992; Stackebrandt & Goebel, 1994). Consequently, near identity of rRNA gene sequences does not eliminate the need to apply other methods to further explore whether isolates are sufficiently similar to be assigned to the same species. Using all to date existing molecular methods, one has always to face an important problem: to decide at what depth of clustering to define species and how to incorporate ecology into species definitions. Many named species demarcations are not rooted in evolutionary or ecological theory, so that many species are extremely diverse in their metabolic capabilities (Feldgarden et al., 2003), in the gene content of their genomes (Welch et al., 2002) and in their ecology (Schloter et al., 2000).
Defining species limits using levels of sequence similarity, typically found within existing named species, is clearly inappropriate. A more attractive approach is to seek ecological, genomic or phenotypic differences among the major clusters resolved by molecular methods that would justify their separation into species. Cohan (2001, 2002, 2004) and Godreuil et al. (2005) have proposed that bacterial species could be split into smaller, more meaningful units by incorporating the concept of the ecotype, and that an “ecotype model” could provide a rational basis for demarcating bacterial taxa. Ecotypes are defined as populations that are genetically cohesive and ecologically distinct. Cohesion results from periodic selection events that recurrently purge each ecotype of its genetic diversity. Moreover, ecotypes are expected to be irreversibly separate from one another. Ecotypes therefore hold all the quintessential properties of species as understood in systematics outside of microbiology (de Queiroz, 1998).
Ecotypes are populations of organisms occupying the same ecological niche, whose divergence is purged recurrently by natural selection. These ecotypes can be discovered by several universal sequence-based approaches. These molecular methods suggest that a typical named species contains many ecotypes, each with the universal attributes of species. A named species is thus likely a genus than a species (Cohan, 2002).
Each ecotype is expected to be identifiable as a sequence cluster, where the average sequence divergence between ecotypes is much greater than the average sequence divergence within them, for any gene shared by the ecotypes. In addition, each ecotype is expected to be identifiable as a monophyletic group in a phylogeny based on DNA sequence data (Cohan, 2002).
The recent studies of morphological variability of isolated strains do not help identification and classification of cyanobacteria. The cultivation is important, but always unifies the culture conditions (which are, moreover, usually stressing for majority of isolates). Therefore, the results about morphology in cultures must be accepted to evaluation of subgeneric units with a special care. The species category is evidently needed in cyanobacteria, but its concept should be probably determined by more diverse and conventional criteria (Castenholz & Norris, 2005; Compere, 2005; Hoffmann, 2005; Johansen & Casamatta, 2005; Komárek et al., 2005; Oren & Tindall, 2005).
The concept of the ecotype provides a rational basis for creating and defining bacterial taxa. In contrary, sequence-derived phylogenies, which organize bacterial diversity into clusters, have certain limitations. First, clustering might not occur in case of continues spectrum of genotypes. Second, it might be impossible to define a new group when a limited number of genotypes have been isolated. Third, phenotypes might not be present by stable chromosomal loci, resulting in marked differences among strains that are closely related. Fourth, in case of new taxa, it might be unclear where to draw the distinction between clusters.
Future prospects of species concept should certainly incorporate ecological data, which will allow real taxonomic assignments.
Molecular phylogeny markers in cyanobacterial classification
Advances in molecular phylogeny in the past decades identified cyanobacteria as the original source of oxygenic photosynthesis and through endosymbiotic incorporation with eukaryotes also the main source of planetary primary production. Cyanobacteria today are associated not only with eutrophication of aquatic habitats, pollution, toxicity, but also with nitrogen fixation in oligotrophic oceans and symbiosis. Yet the genetic identity and specific ecological roles are only started to be explored, although the molecular tools are now largely available.
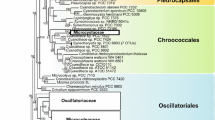
Advances in molecular phylogeny (Woese & Fox, 1977; Woese, 1987) revealed the significance of cyanobacteria as the monophyletic origin of oxygenic photosynthesis. The first reconstruction of the phylogenetic interrelationships among cyanobacteria (Giovannoni et al., 1988) was based on 16S rRNA of organisms maintained in axenic cultures. It provided an insight in early diversification of the group. This scheme has been compared with morphotypic expression of the examined taxa and found some encouraging correlations, but identified also the polyphyletic nature of some traditional botanically as well as bacteriologically established genera (Willmote, 1994). Since then, the GenBank has been enriched with numerous complete and partial sequences of the 16S rRNA gene derived from axenic and non-axenic uni-cyanobacterial cultures, as well as from natural populations. This data provides a useful matrix within which the overall phylogenetic relations were reconstructed (Rudi et al., 1997; Willmote & Herdman, 2001).
Further refinement in resolution of closer phylogenetic relations was achieved by comparing spacer sequences (ITS) between 16S rRNA and 23S rRNA genes (Iteman et al., 2000). Characterization of the ability and evolution of nitrogen fixation in cyanobacteria by identification and sequencing of the nifH gene were a particular success (Stewart, 1985; Zehr et al., 2003). Comparison of nifH and ribosomal RNA phylogenies from cultivated microorganisms showed no conclusive evidence of widespread lateral gene transfer, thereby further supporting the potential for a phylogenetic basis of future cyanobacterial systematics.
With the application of cyanobacterial-specific primers (Urbach et al., 1992; Nübel et al., 1997; Laloui et al., 2002), research on cyanobacterial natural populations has been applied with increasing success, joining other culture-independent methods in microbial ecology (e.g. Amann et al., 1995). Culture-independent analysis of sequences derived from samples of environmental genomic nucleic acids has revolutionized our understanding of cyanobacterial diversity, function and processes (Stahl et al., 1984; Hugenholtz & Pace, 1996; Bates et al., 2012; Steven et al., 2012). Technological advances such as e.g. pyrosequencing enable rapid characterization of cyanobacterial communities that are faster and at greater sequence depth than was deemed possible via cloning and Sanger sequencing (Sogin et al., 2006).
Since the early days of a bacteriological approach to cyanobacterial taxonomy, however, Krumbein (1979) and Rippka et al. (1979) have pointed to the importance of both traditional and molecular approaches. Golubic (1979) has stressed the importance of the taxonomic hyperspace or multidimensional correlation scheme in numerical taxonomy according to Sneath & Sokal (1973).
There are several examples showing the clear necessity of applying polyphasic approach in taxonomy of cyanobacteria (Castenholz, 1992; Palinska et al., 1996; Otsuka et al., 2000; Lyra et al., 2005; Rajaniemi et al., 2005). Palinska et al. (1996) indicated in their studies on Synechococcus/Synechocystis/Merismopedia/Eucapsis complex that the great morphological diversity observed in nature and (partially) in culture does not necessarily reflect genetic diversity. In fact, much less cyanobacterial species diversity seems to exist in culture and probably also in nature than have been described according to the morphological features of the Botanical Code. Cyanobacterial diversity should certainly be based not only on genetic similarity of the 16S rRNA gene but also on similarities of different gene fragments . Phenotypic and phylogenetic analyses on 16S rRNA gene fragments done by Garcia-Pichel et al. (1996) have shown that Microcoleus chthonoplastes is a cosmopolitan cyanobacterium. Interestingly, Lodders et al. (2005) could provide the contrary, data on rRNA-ITS locus showed that this species sharing similar morphology differ on the genetic level. Similarly, Otsuka et al. (2001) using the cpcBA intergenic spacer and 16S–23S internal transcribed spacer concluded that the six Microcystis morphospecies: M. aeruginosa, M. ichthyoblabe, M. novacekii, M. viridis, M. wesenbergii, M. flos-aquae and M. pseudofilamentosa, may possibly be unified into one species.
Traditional phenotypic properties (morphological and physiological features) such as development (type of reproduction and division pattern), structure (cell size and shape or type of trichome, arrangement of cells, heterocyst and akinete formation, motility), and physiology (e.g. chromatic adaptations, salinity tolerance, vitamin requirements) were proved to be variable with changing environmental and culture conditions (Rippka et al., 1979; Dor & Hornhoff, 1985; Holtkamp, 1985; Castenholz & Waterbury, 1989; Palinska et al., 1996; Otsuka et al., 2000; Lyra et al., 2005; Rajaniemi et al., 2005).
In recent years, a number of valuable phenotypic, especially ultramorphological features, have been confirmed to be stable and reliable taxonomic characters (Komárek & Anagnostidis 1999, 2005). Palinska et al. (1998) and Palinska & Krumbein (2000) emphasized the taxonomic value of cell wall perforations. Their number, size and organization were studied in nine species belonging to different genera and have been proved to be stable and similar in different life stages of organisms and under different laboratory conditions. However, the most important feature of inner cell structures which are usable for taxonomic classification is thylakoids. Their arrangement is supposed to be uniform in all studied orders and families (Hernandez-Marine & Wit, 1999; Komárek & Anagnostidis 1999, 2005). However, the results of Marquardt & Palinska (2007) and Palinska & Marquardt (2008) cannot support this statement. Strains belonging to the morphotype Phormidium autumnale and other Phormidium species showed definitely divergent thylakoids scheme.
The current taxonomy of cyanobacteria still depends too much upon morphological characteristics and must be reviewed by means of bacteriological methods as well as traditional botanical methods. Molecular investigations, especially those based on 16S rRNA and DDH, would bring unification or division to some species, genera and even taxa of higher rank. There remain many cyanobacteria to be reconsidered regarding their taxonomy. The genus Synechococcus is an example; genetic distances among Synechococcus spp. in the neighbour-joining tree are too large for them to be classified in a single genus, thus it has been suggested that this is another example of an unnaturally large taxon (Honda et al., 1999).
Similar situation is faced in case of the representatives of the genus Phormidium. Cyanobacteria included into Phormidium-like group occur in an enormous diversity. Observations on morphologically complex cyanobacteria in freshwater, marine and terrestrial environments (Turner, 1997; Wilmotte & Herdman, 2001; Marquardt & Palinska, 2007) showed that the distribution of populations of these morphologically complex cyanobacteria follows patterns that correlate rather with ecological determinants than with the organisms determined as “Phormidium”. The results of the multiple 16S rRNA, ITS and phycocyanin intergenic spacer (cpcBA-IGS) sequence analyses (Marquardt & Palinska, 2007) revealed that Phormidium group is not phylogenetically coherent and has demonstrated the need for drastic revisions of this group in the future (Wilmotte & Herdman, 2001).
The only acceptable and recommendable method for modern taxonomic evaluation of cyanobacteria is the combined approach with use of phenotype, ultrastructural, ecological, biochemical and molecular methods.
Herbarium specimens as a source of molecular records potentially harmonizing the classical and the modern cyanobacterial taxonomies
Microbiologists and botanists studying taxonomy or phylogeny of cyanobacteria should not assume that the strains they have used have been correctly identified (Wilmotte & Herdman, 2001), and that many culture collections contain misidentified strains. One should be aware that the various “phylogenetic” trees illustrating similarities based on sequences from particular parts of the genome often use results from strains whose generic and specific names are doubtful. In most cases, these strains have been maintained in culture for decades, they have lost morphological and physiological properties of determinative value and many are of unknown origin. This restricts the value of such trees and analyses for comparing possible evolutionary relationships (Whitton & Potts, 2000). The reasons for that are i) difficulties in morphological identification, ii) few characterized strains available and iii) no verification of strain identification.
Palinska et al. (2006) in the course of their study on diversity and taxonomy of cyanobacteria decided to start molecular research also on historical and dried environmental samples of cyanobacteria. The polyphasic characterization and identification of historical strains from exsiccate were compared to the traditional lists of strains established on classical, morphological criteria by Rabenhorst (1873), Drouet and Gomont.
The triggers for this type of studies were as follows:
-
Existence of two independent Codes of Nomenclature in case of cyanobacteria;
-
The number of names of cyanobacterial species that have been validly published under the Bacteriological Code is extremely small. No more than five genera (Halospirulina, Planktotricoides, Prochlorococcus, Prochloron and Prochlorothrix) and 13 names of cyanobacterial species have been proposed so far in the International Journal of Systematic and Evolutionary Microbiology (IJSEM)/International Journal of Systematic Bacteriology (IJSB);
-
Only five species names (Halospirulina tapeticola, Prochlorococcus marinus, Prochloron didemni, Prochlorothrix hollandica and Planktotricoides raciborskii) out of 13 are validly published (Oren, 2004);
-
Lack of clear species definition and description for cyanobacteria;
-
Urgent need of uniform "type species" for botanists and bacteriologists;
-
Cyanophytes “type species” has never before been explored in terms of its genetic properties;
-
Molecular methods and morphological data provide a new scaffold for the accumulated taxonomic knowledge on historical cyanobacterial herbaria.
The advantage of using botanical-type material from herbaria in molecular approaches is that these specimens have complete, proved and accepted strain history and morphological description. Therefore, 16S rRNA sequences from herbarium specimens can be used as references in various phylogenetic and taxonomic relationships. Morphology of the specimens used had been carefully described already more than 100 years ago and confirmed using different microscopical approaches. Herbarium collections are perfect objects for intensive phylogenetic studies, although they have not been used for these purposes previously. We strongly encourage scientists interested in phylogeny of cyanobacteria to use the morphological data and descriptions present in exsiccate collections for their studies. Correct phenotypic descriptions would enormously help the interpretation of phylogenetic trees and avoid the misleading data derived from studies that examine only accidentally isolated and misidentified organisms.
Thanks to genetic studies on herbarium samples, botanical-type specimens have been for the first time explored and characterized in terms of their genetic as well as phenotypic properties. This opened the possibility to validate a large volume of ecologically relevant research and connect the modern polyphasic assessment of diversity with traditional phenotype-based identifications and floral listings. It was tested for the first time whether microorganisms identified by phenotypic characters indeed correspond to the same genotype as traditionally assumed. That research confirmed the designation of each studied herbarium sample by phylogenetic analyses (Palinska et al., 2006).
Further polyphasic studies on exsiccate would help in answering the question whether the phycological practice of relying on fixed-type specimens for reference or the bacteriological one using live axenic cultures as type reference is more reliable. The hypothesis of phycologists is that axenic-type cultures evolve, thus the reference changes over time. The argument against phycological practice is that phenotypes may be expressions of different genotypes, or be entirely controlled by environment. Furthermore molecular comparison of present toxic or e.g. nitrogen fixing strains, with the morphologically identical type-material should be performed in order to test the genotypic changes over time.
The historic collections contain precious information and should be maintained since they are amenable to new technologies and molecular approaches and enable inferences to be made about historic populations.
Scientists interested in the phylogeny of cyanobacteria are encouraged to consult the morphological data and descriptions present in collections of exsiccata for their studies as correct phenotypic descriptions would enormously help the interpretation of phylogenetic trees and avoid misleading information derived from studies that examine only accidentally isolated and misidentified organisms.
Molecular data gained for the botanical-type specimens allow doing a first step in the unification of the two Bacteriological and Botanical Codes, in the case of cyanobacteria. Thanks to modern molecular data, botanical-type species received genetic definition required by both codes.
References
Amann, R. I., W. Ludwig & K.-H. Schleifer, 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews 59: 143–169.
Anagnostidis, K. & J. Komárek, 1985. Modern approach to the classification system of cyanophytes, Introduction. Archiv für Hydrobiologie. Supplementband 71. Algological Studies 38(39): 291–302.
Bates, S. T., D. Berg-Lyons, C. L. Lauber, W. A. Walters, R. Knight & N. Fiefer, 2012. A preliminary survey of lichen associated eukaryotes using pyrosequencing. The Lichenologist 44: 137–146.
Bornet, E. & C. Flahaut, 1887. Révision des Nostocacées hétérocystées. Annales des Sciences Naturelles (Botanique) 5: 51–129.
Bornet, E. & C. Flahaut, 1888a. Note sur deux nouveaux genres d’algues perforantes. Journal de Botanique 2: 161–165.
Bornet, E. & C. Flahaut, 1888b. Revision des Nostocacées hétérocystées. Annales des Sciences Naturelles (Botanique) 7: 177–262.
Bourrelly, P., 1970. Les algues d’eau douce III. Boubée & Cie, Paris.
Castenholz, R. W., 1992. Species usage, concept, and evolution in the cyanobacteria (blue-green algae). Journal of Phycology 28: 737–745.
Castenholz, R. W., 2001. General characteristics of cyanobacteria. In Garrity, G. M. (ed.), Bergey’s Manual of Systematic Bacteriology. Springer, New York: 474–478.
Castenholz, R. W. & T. B. Norris, 2005. Revisionary concepts of species in the cyanobacteria and their applications. Algological Studies 117: 53–69.
Castenholz, R. W. & J. B. Waterbury, 1989. Group I. Cyanobacteria. Preface. In Holt, J. G. (ed.), Bergey´s Manual of Systematic Bacteriology, Vol. 3. Williams & Wilkins, Baltimore: 1710–1727.
Cohan, F. M., 2001. Bacterial species and speciation. Systematics Biology 50: 513–524.
Cohan, F. M., 2002. What are bacterial species? Annual Review of Microbiology 56: 457–487.
Cohan, F. M., 2004. Concepts of bacterial biodiversity for the age of genomics. In Fraser, C. M., T. Read & K. E. Nelson (eds), Microbial Genomes. Humana Press, New York: 175–194.
Compere, P., 2005. The nomenclature of the Cyanophyta under the Botanical Code. Algological Studies 117: 31–37.
Dawson, C. A. & P. H. A. Sneath, 1985. A probability matrix for the identification of vibrios. Journal of Applied Bacteriology 58: 407–423.
de Queiroz, K., 1998. Species and speciation. In Howard, D. J. & S. H. Berlocher (eds), Endless Forms. Oxford University Press, Oxford: 57–75.
Desikachary, T. V., 1959. Cyanophyta. Indian Council Agricultural Research. Academic Press, New York.
Dor, I. & M. Hornhoff, 1985. Salinity-temperature relations and morphotypes of mixed population of coccoid cyanobacteria from a hot, hypersaline pond in Israel. Marine Ecology 6: 13–25.
Drouet, F., 1981. Summary of the classification of blue-green algae. Nova Hedwigia, Beihefte 66: 133–209.
Elenkin, A. A., 1938 & 1949. Sinezelenye vodorosli SSSR Monographia algarum cyanophycearum aquidulcium et terrestrium in finibus URSS inventarum. Pars spec 1, 2, Akademia Nauk SSSR, Moscow-Leningrad.
Feldgarden, M., N. Byrd & F. M. Cohan, 2003. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology 149: 3565–3573.
Fox, G. E., J. D. Wisotzkey & P. Jurtshuk, 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. International Journal of Systematic Bacteriology 42: 166–170.
Friedmann, E. I. & L. Borowitzka, 1982. The symposium on taxonomic concepts in blue-green algae: towards a compromise with the bacteriological code? Taxon 31: 673–683.
Fritsch, F. E., 1959. The Structure and Reproduction of the Algae, Vol. 1. University Press, Cambridge.
Garcia-Pichel, F., L. Prufert-Bebout & G. Muyzer, 1996. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Applied and Environmental Microbiology 62: 3284–3291.
Geitler, L., 1932. Cyanophyceae. In Kolkwitz, R. (ed.), Rabenhorst‘s Kryptogamen Flora von Deutschland, Österreich und der Schweiz, Vol. 14. Akademische Verlagsgesellschaft, Leipzig: 1–1196.
Giovannoni, S. J., S. Turner, G. J. Olsen, J. Barns, D. J. Lane & N. R. Pace, 1988. Evolutionary relationships among cyanobacteria and green chloroplasts. Journal of Bacteriology 170: 3584–3592.
Godreuil, S., F. M. Cohan, H. Shah & M. Tibayrenc, 2005. Which species concept for pathogenic bacteria? An e-debate. Infection, Genetics and Evolution 5: 375–387.
Golubic, S., 1976. Organisms that build stromatolites. In Walter, M. R. (ed.), Stromatolites, Developments in Sedimentology. Elsevier Scientific Publishing Co., Amsterdam: 113–126.
Golubic, S., 1979. Einführung in die Probleme der numerischen Taxonomie bei Cyanophyten. In Krumbein, W. E. (ed.), Cyanobacterien oder Algen?. Oldenburg, Littman: 15–32.
Gomont, M., 1892. Monographie des Oscillariées (Nostocaceae homocystées). Annales des Sciences Naturelles, Serie Botanique 15: 265–368.
Hernandez-Marine, M. & R. Wit, 1999. Characterization and morphological transformation of laboratory grown Leptolyngbya valderiana (Gom.) Anagn. et Komárek (Cyanophyta/Cyanobacteria). Algological Studies 94: 139–146.
Hoffmann, L., 2005. Nomenclature of Cyanophyta/Cyanobacteria: roundtable on the unification of the nomenclature under the Botanical and Bacteriological Codes. Algological Studies 117: 13–29.
Holtkamp, E., 1985. The microbial mats of the Gavish Sabkha (Sinai). Environmental factors as determinants of the formation, structure and composition of hypersaline cyanobacterial associations. PhD Thesis University of Oldenburg, Germany.
Honda, D., A. Yokota & J. Sugiyama, 1999. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. Journal of Molecular Evolution 48: 723–739.
Hugenholtz, P. & N. R. Pace, 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends in Biotechnology 14: 190–197.
Iteman, I., R. Rippka, N. Tandeau de Marsac & M. Herdman, 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer of cyanobacteria. Microbiology 146: 1275–1286.
Johansen, J. R. & D. A. Casamatta, 2005. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algological Studies 117: 71–93.
Komárek, J., 2013. Cyanoprokaryota 3. Teil Heterocystous Genera. In Büdel, B., G. Gärtner, L. Krienitz & M. Schagerl (eds), Süßwasserflora von Mitteleuropa. Spektrum Akademischer Verlag.
Komárek, J. & K. Anagnostidis, 1999. Cyanoprokaryota 1. Teil Chroococcales. In Ettl, H., G. Gärtner, H. Heynig & D. Mollenhauer (eds), Süßwasserflora von Mitteleuropa. Gustav Fischer, Jena-Stuttgart-Lübeck-Ulm.
Komárek, J. & K. Anagnostidis, 2005. Cyanoprokaryota 2. Teil Oscillatoriales. In Büdel, B., G. Gärtner, L. Krienitz & M. Schagerl (eds), Süßwasserflora von Mitteleuropa. Gustav Fischer, Jena-Stuttgart-Lübeck-Ulm.
Komárek, J., S. Ventura, S. Turicchia, J. Komarkova, C. Mascalchi & E. Soldati, 2005. Cyanobacterial diversity in alkaline marshes of northern Belize (Central America). Algological Studies 117: 265–278.
Kondrateva, N. V., 1968. Sino-zeleni vodorosti. In Vizn. Prisnov. Vodorost. Ukr. RSR 1, 2, Naukova Dumka, Kiev.
Krumbein, W. E., 1979. Über die Zuordnung der Cyanophyten. In Krumbein, W. E. (ed.), Cyanobacterien oder Algen?. Oldenburg, Littman: 33–48.
Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau de Marsac, M. Herdman & I. Iteman, 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the “marine Synechococcus clade” by size, sequence analyses or RFLP of the internal transcribed spacer (ITS) of the ribosomal operon. Microbiology 148: 453–455.
Lodders, N., E. Stackebrandt & U. Nübel, 2005. Frequent genetic recombination in natural populations of the marine cyanobacterium Microcoleus chthonoplastes. Environmental Microbiology 7: 434–442.
Lyra, C., M. Laamanen, J. Lehtimäki, A. Surakka & K. Sivonen, 2005. Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. International Journal of Systematic and Evolutionary Microbiology 55: 555–568.
Marquardt, J. & K. A. Palinska, 2007. Genotypic and phenotypic diversity of cyanobacteria assigned to the genus Phormidium (Oscillatoriales) from different habitats and geographical sites. Archives of Microbiology 187: 397–413.
Mayr, E., 1982. The Growth of the Biological Thought. The Belknap Press, Cambridge.
Moore, L., R. G. Rocap & S. W. Chisholm, 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393: 464–467.
Nübel, U., F. Garcia-Pichel & G. Muyzer, 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Applied and Environmental Microbiology 63: 3327–3332.
Oren, A., 2004. A proposal for further integration of the cyanobacteria under the Bacteriological Code. International Journal of Systematic and Evolutionary Microbiology 54: 1895–1902.
Oren, A., 2011. Naming Cyanophyta/Cyanobacteria – a bacteriologist’s view. Fottea 11: 9–16.
Oren, A., 2014. Cyanobacteria: biology, ecology and evolution. In Sharma, N. K., A. K. Rai & L. Stal (eds) Stress Biology of Cyanobacteria: Molecular Mechanisms to Cellular Responses. Wiley, New York: 3–20.
Oren, A. & G. M. Garrity, 2014. Proposal to change general consideration 5 and principle 2 of the International Code of Nomenclature of Prokaryotes. International Journal of Systematic and Evolutionary Microbiology 64: 309–310.
Oren, A. & B. J. Tindall, 2005. Nomenclature of the Cyanophyta/Cyanobacteria/Cyanoprokaryotes under the International Code of Nomenclature of Prokaryotes. Algological Studies 117: 39–52.
Otsuka, S., S. Suda, R. Li, S. Matsumoto & M. M. Watanabe, 2000. Morphological variability of colonies of Microcystis morphospecies in culture. Journal of General and Applied Microbiology 46: 57–68.
Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto & M. M. Watanabe, 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kützing ex Lemmermann 1907 under the Rules of the Bacteriological Code. International Journal of Systematic and Evolutionary Microbiology 51: 873–879.
Palinska, K. A. & W. E. Krumbein, 2000. Perforation patterns in filamentous cyanobacteria. Journal of Phycology 36: 139–145.
Palinska, K. A., W. E. Krumbein & U. Schlemminger, 1998. Ultramorphological studies on Spirulina sp. Botanica Marina 41: 349–355.
Palinska, K. A., W. Liesack, E. Rhiel & W. E. Krumbein, 1996. Phenotype variability of identical genotypes: the need for a combined cyanobacterial taxonomy demonstrated on Merismopedia-like isolates. Archives of Microbiology 166: 224–233.
Palinska, K. A. & J. Marquardt, 2008. Conformity between molecular and phenotypic results demonstrated on Phormidium autumnale isolates (cyanobacteria). Archives of Microbiology 189: 325–335.
Palinska, K. A. & W. Surosz, 2008. Population of Aphanizomenon from the Gulf of Gdansk (Southern Baltic Sea): differences in phenotypic and genotypic characteristics. Hydrobiologia 607: 163–173.
Palinska, K. A., C. F. Thomasius, J. Marquardt & S. Golubic, 2006. Phylogenetic evaluation of cyanobacteria preserved as historic herbarium exsiccata. International Journal of Systematic and Evolutionary Microbiology 56: 2253–2263.
Rabenhorst, L., 1873. Algarum europæarum exsiccatarum : (der Algen Europa’s mit Berücksichtigung des ganzen Erdballs). Verlag Dresden Drück von C. Heinrich: 1–235.
Rajaniemi, R., P. Hrouzek, K. Kastovska, R. Willame, A. Rantala, L. Hoffmann, J. Komarek & K. Sivonen, 2005. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria). International Journal of Systematic and Evolutionary Microbiology 55: 11–26.
Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman & R. Y. Stanier, 1979. Generic assignments, strain histories and properties of pure culture of cyanobacteria. Journal of General Microbiology 111: 1–61.
Rippka, R., T. Coursin, W. Hess, C. Lichtlé, D. J. Scanlan, K. A. Palinska, I. Iteman, F. Partensky, J. Houmard & M. Herdman, 2000. Prochlorococcus marinus. Chisholm et al. 1992, subsp. nov. pastoris, strain PCC 9511, the first axenic chlorophyll a2b2 marine, chlorophyll b-containing cyanobacterium (Oxyphotobacteria). International Journal of Systematic and Evolutionary Microbiology 50: 1833–1847.
Rudi, K., O. M. Skulberg, F. Larsen & K. S. Jakobsen, 1997. Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7, and V8. Applied and Environment Microbiology 63: 2593–2599.
Schloter, M., M. Lebuhn, T. Heulin & A. Hartmann, 2000. Ecology and evolution of bacterial microdiversity. FEMS Microbiology Review 24: 647–660.
Sneath, P. H. A. & R. R. Sokal, 1973. Numerical Taxonomy. Freemann, San Francisco: 1–573.
Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta & G. J. Herndl, 2006. Microbial diversity in the deep sea and the under explored ‘‘rare biosphere’’. Proceedings of the National Academy of Sciences 103: 12115–12120.
Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward & W. B. Whitman, 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology 52: 1043–1047.
Stackebrandt, E. & B. M. Goebel, 1994. A place for DNA–DNA reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic Bacteriology 44: 846–849.
Stahl, D. A., D. J. Lane, G. J. Olsen & N. R. Pace, 1984. Analysis of hydrothermal vent-associated symbionts by ribosomal-RNA sequences. Science 224: 409–411.
Stanier, R. Y., W. R. Sistrom, T. A. Hansen, B. A. Whitton, R. W. Castenholz, N. Pfennig, V. N. Gorlenko, E. N. Kondratieva, K. E. Eimhjellen, R. Whittenbury, R. L. Gherna & H. G. Trüper, 1978. Proposal to place the nomenclature of the cyanobacteria (blue-green algae) under the rules of the International Code of Nomenclature of Bacteria. International Journal of Systematic Bacteriology 28: 335–336.
Starmach, K., 1966. Cyanophyta, Sinice. Flora Slodkowodna Polski, Vol. 2. Polska Akademia Nauk, Warszawa: 1–753.
Steven, B., S. McCann & N. L. Ward, 2012. Pyrosequencing of plastid 23S rRNA genes reveals diverse and dynamic cyanobacterial and algal populations in two eutrophic lakes. FEMS Microbiology Ecology 82: 607–615.
Stewart, W. D. P., 1985. Nitrogen fixation: differentiating cyanobacteria rearrange their nif genes. Nature 314: 404–405.
Thuret, G., 1875. Essai de classification des Nostochinées. Annales des Sciences Naturelles (Botanique) 6: 372–382.
Turner, S., 1997. Molecular systematics of oxygenic photosynthetic bacteria. In Bhattacharya, D. (ed.), The Origin of the Algae and their Plastids. Springer, New York: 13–52.
Urbach, E., D. Robertson & S. W. Chisholm, 1992. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355: 267–269.
Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters & J. Swings, 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiological Reviews 60: 407–438.
Waterbury, J. B. & R. Y. Stanier, 1977. Two unicellular cyanobacteria which reproduce by budding. Archives of Microbiology 115: 249–257.
Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg & F. R. Blattner, 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proceedings of the National Academy of Sciences 99: 17020–17024.
Whitton, B. A. & M. Potts, 2000. Introduction to the cyanobacteria. In Whitton, B. A. & M. Potts (eds), The Ecology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht: 1–11.
Wilmotte, A., 1994. Molecular evolution and taxonomy of the cyanobacteria. In Bryant, D. A. (ed.), The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht: 1–25.
Wilmotte, A. & M. Herdman, 2001. Phylogenetic relationships among the cyanobacteria based on 16S rRNA sequences. In Garrity, G. M. (ed.), Bergey’s Manual of Systematic Bacteriology: 487–493.
Woese, C. R., 1987. Bacterial evolution. Microbiological Reviews 51: 221–271.
Woese, C. R. & G. E. Fox, 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences 74: 5088–5090.
Zehr, J. P., B. D. Jenkins, S. M. Short & G. F. Steward, 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environmental Microbiology 5: 539–554.
Acknowledgments
This work was supported by DFG grant Pa 842/9-1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Stefano Amalfitano
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Palinska, K.A., Surosz, W. Taxonomy of cyanobacteria: a contribution to consensus approach. Hydrobiologia 740, 1–11 (2014). https://doi.org/10.1007/s10750-014-1971-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-014-1971-9