Abstract
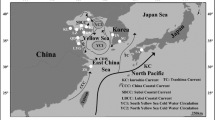
Portunus trituberculatus is a commercially important species widely spread in the East China Sea. Intraspecific variation of the mitochondrial DNA cytochrome oxidase subunit I (mtDNA COI) gene was investigated in 213 individuals from six localities (Changjiang Estuary, Shengsi Islands, Zhoushan Islands, Dongtou Islands, Dinghai Bay, and Quanzhou Bay) ranging from north (31°21′N) to south (24°55′N) coastal waters of the East China Sea. Overall, a total of 27 mtDNA haplotypes and 21 variable sites were detected in the 787 bp segment of COI gene. Analysis of mtDNA COI sequence data revealed that crabs from the six localities were characterized by moderately high haplotypic diversity (h = 0.787 ± 0.026), while sequence divergence values between haplotypes were relatively low (π = 0.00241 ± 0.00098). Each population was characterized by a single most frequent haplotype, shared among all six localities, and a small number of rare ones, typically present in only one or two individuals and representative of a specific population. However, neither the neighbor-joining tree nor the minimum spanning network (MSN) based on the haplotype data exhibited geographical patterns of the six populations. Mismatch distribution analysis of P. trituberculatus individuals sampled from the six localities suggested that sudden population expansion might have occurred in CJ and SS population that might be consistent with over-exploitation of the swimming crab. Analysis of molecular variance (AMOVA) and F ST statistics showed that significant genetic differentiation existed among the SS, ZS, DT, DH, and QZ populations, suggesting that gene flow might be reduced, even between the geographically close sites, despite the high potential of dispersal. The possible causes of the observed genetic heterogeneity among the P. trituberculatus populations and the potential applications of the mtDNA COI marker in the artificial breeding and fisheries management are discussed.



Similar content being viewed by others
References
Avise, J. C., 1987. Identification and interpretation of mitochondrial DNA stocks in marine species. In Kumpf, H. (ed.), Proceedings of the Stock Identification Workshop. U.S. Department of Commerce NOAA Technical Memorandum NMFS-SEFC-199: 105–136.
Avise, J. C., 2000. Phylogeography. Harvard University Press, London: 447 pp.
Azuma, N., Y. Kunihiro, J. Sasaki, E. Mihara, Y. Mihara, T. Yasunaga, D. H. Jin & S. Abe, 2008. Genetic variation and population structure of hair crab (Erimacrus isenbeckii) in Japan inferred from mitochondrial DNA sequence analysis. Marine Biotechnology 10: 39–48.
Cassone, B. J. & E. G. Boulding, 2006. Genetic structure and phylogeography of the lined shore crab, Pachygrapsus crassipes, along the northeastern and western Pacificcoasts. Marine Biology 149: 213–226.
Cho, E. S., C. G. Jung, S. G. Sohn, C. W. Kim & S. J. Han, 2007. Population genetic structure of the ark shell Scapharca broughtonii schrenck from Korea, China, and Russia based on COI gene sequences. Marine Biotechnology 9: 203–216.
Dai, A. Y., 1977. Primary investigation on the fishery biology of the Portunus trituberculatus. Marine Fisheries 25: 136–141 (in Chinese).
Dai, A. Y., S. L. Yang & Y. Z. Song, 1986. Marine crabs in China Sea. Marine Publishing Company, Beijing: 194–196 (in Chinese).
Excoffier, L., P. E. Smouse & J. M. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 491–497.
Excoffier, L. & P. E. Smouse, 1994. Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136: 343–359.
Excoffier, L., G. Laval & S. Schneider, 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Fu, Y. X., 1997. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics 147: 915–925.
Gao, B. Q., P. Liu, J. Li & F. Y. Dai, 2007. Isozyme polymorphism in Portunus trituberculatus from wild population. Journal of Fisheries of China 31: 1–6 (in Chinese).
Harpending, H., 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology 66: 591–600.
Harpending, H., S. T. Sherry, A. R. Rogers & M. Stoneking, 1993. The genetic structure of ancient human populations. Current Anthropology 34: 483–496.
Hedgecock, D., 1986. Is gene flow from pelagic larval dispersal important in the adaptation and evolution of marine invertebrates? Bulletin of Marine Science 39: 550–564.
Hedgecock, D., 1994. Temporal and spatial genetic structure of marine animal populations in the California current. California Cooperative Oceanic Fish Invest Reports 35: 73–81.
Imai, H., Y. Fujii, J. Karakawa, S. Yamamoto & K. Numachi, 1999. Analysis of the population structure of the swimming crab, Portunus trituberculatus in the coastal waters of Okayama Prefecture, by RFLPs in the whole region of mitochondrial DNA. Fisheries Science 65: 655–656.
Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequence. Journal of Molecular Evolution 16: 111–120.
Kumar, S., K. Tamura & M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150–163.
Lavery, S., C. Moritz & D. R. Fielder, 1996. Indo-Pacific population structure and evolutionary history of the coconut crab Birgus latro. Molecular Ecology 5: 557–570.
Li, P. F., P. Liu, J. Li, F. Y. Dai & Y. Y. He, 2007. Biochemical genetic analysis of Portunus trituberculatus in Laizhou Bay. Marine Fisheries Research 28: 90–96 (in Chinese).
Lintas, C., J. Hirano & S. Archer, 1998. Genetic variation of the European eel (Anguilla anguilla). Molecular Marine Biology and Biotechnology 7: 263–269.
Maes, G. E. & F. A. M. Volckaert, 2002. Clinal genetic variation and isolation by distance in the European eel Anguilla anguilla (L.). Biological Journal of the Linnean Society 77: 509–521.
Palumbi, S. R., 1992. Marine speciation on a small planet. Trends in Ecology & Evolution 7: 114–118.
Palumbi, S. R., 1994. Genetic divergence, reproductive isolation and marine speciation. Annual Review of Ecology and Systematics 25: 547–572.
Perrin, C., S. R. Wing & M. S. Roy, 2004. Effects of hydrographic barriers on population genetic structure of the sea star Coscinasterias muricata (Echinodermata, Asteroidea) in the New Zealand fiords. Molecular Ecology 13: 2183–2195.
Rogers, A. R., 1995. Genetic evidence for a Pleistocene population explosion. Evolution 49: 608–615.
Rogers, A. R. & H. Harpending, 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology Evolution 9: 552–569.
Roman, J. & S. R. Palumbi, 2004. A global invader at home: population structure of the green crab, Carcinus maenas, in Europe. Molecular Ecology 13: 2891–2898.
Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer & R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Saitou, N. & M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology Evolution 4: 406–425.
Sambrook, J., E. F. Fritsh & T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York.
Schneider, S. & L. Excoffier, 1999. Estimation of past demographic parameters from the distribution of pairwise differences when mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089.
Shen, J. R. & R.Y. Liu, 1965. Shrimps and Crabs in China. Science Popularization Press, Beijing: 23 pp.
Slatkin, M., 1993. Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47: 264–279.
Slatkin, M. & R. R. Hudson, 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562.
Song, H. T., Y. P. Ding & Y. J. Xu, 1989. Population component characteristics and migration distribution of the Portunus trituberculatus in the coast water of Zhejiang Province. Marine Bulletin 8: 66–74 (in Chinese).
Sun, Y. M., Y. Yan & J. J. Sun, 1984. Larvae development of the swimming crab Portunus trituberculatus. Journal of Fisheries of China 8: 219–226 (in Chinese).
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Tajima, F. & M. Nei, 1984. Estimation of evolutionary distance between nucleotide sequences. Molecular Biology Evolution 1: 269–285.
Tave, D., 1993. Genetics for Fish Hatchery Managers. Van Nostrand Reinhold, New York: 415 pp.
Thompson, J. D., D. G. Higgins & J. J. Gibson, 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680.
Wang, G. L., S. Jin, Z. Li & Y. E. Chen, 2005. Tissue specificity and biochemical genetic analysis of isozyme on cultured Portunus trituberculatus stock. Journal of Oceanography in Taiwan Strait 24: 474–480 (in Chinese).
Wirth, T. & L. Bernatchez, 2001. Genetic evidence against panmixia in the European eel. Nature 409: 1037–1040.
Wu, C. W., S. C. Yu & Y. L. Lv, 1996. Fishery Techniques of the Swimming Crab Portunus trituberculatus. Shanghai Science and Technology Press, Shanghai: 28–31 (in Chinese).
Xue, J. Z., N. S. Du & W. Nai, 1997. The researches on the Portunus trituberculatus in China. The East China Sea 15: 60–64 (in Chinese).
Yamauchi, M. M., M. U. Miya & M. Nishida, 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene 311: 129–135.
Yu, C. G., H. T. Song & G. Z. Yao, 2003. Geographical distribution and faunal analysis of crab resources in the East China Sea. Journal of Zhejiang Ocean University 22: 108–113 (in Chinese).
Yu, C. G., H. T. Song & G. Z. Yao, 2004. Assessment of the crab stock biomass in the continental shelf waters of the East China Sea. Journal of Fisheries of China 28: 41–46 (in Chinese).
Acknowledgments
We would like to express our thanks to all the people in Lianjiang Fisheries and Management Bureau for help in sample collection. This work is supported in part by grants from the Shanghai Leading Academic Discipline Project (T1101), the Shanghai Marine Bio-resources and Environment Innovation Platform Project, and from the Shanghai 908 Project (Synthetical Investigations and Evaluations on Shanghai Coastal Waters) (PJ1-2, ST1/ST2).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: C. Sturmbauer
Rights and permissions
About this article
Cite this article
Liu, Y., Liu, R., Ye, L. et al. Genetic differentiation between populations of swimming crab Portunus trituberculatus along the coastal waters of the East China Sea. Hydrobiologia 618, 125–137 (2009). https://doi.org/10.1007/s10750-008-9570-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-008-9570-2