Abstract
The effects of culture media on DNA methylation process, which is one of the epigenetic mechanisms, have not been clearly elucidated although it is known that in vitro culture conditions alter epigenetic mechanisms. This study was designed to address the question: does embryo culture media approach, sequential or single step, differentially affect DNA methylating enzymes and global DNA methylation. Mouse zygotes were cultured either in single step or sequential culture media until the blastocyst stage and in vivo developed blastocyst were utilized as control. Similarly, GV stage oocytes were in vitro matured either in single step or first step of sequential culture media. In vivo matured MII oocytes were used as control. The expression levels and cellular localization of Dnmt1 and 3a enzymes were analyzed by immunofluorescence and western blot analysis while global DNA methylation was evaluated by immunofluorescence. We found that signal intensities of Dnmt1 and Dnmt3a enzymes were significantly low in embryos or oocytes cultured in sequential media compared to single step media and control, which were comparable amongst themself. Similarly, global DNA methylation level in single step media and control groups was comparable but both was higher than the sequential media. This study demonstrated that composition of culture media may differentially affect DNA methylation levels in mouse embryos and oocytes. Since abnormal DNA methylation may cause aberrant oocyte or embryo development, we think that further studies are needed to test human embryos and oocyte, and to explain molecular mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human assisted reproduction technologies (ARTs) have been implemented in the treatment of infertility during the four decades. As ARTs involve several steps subjecting the gametes and early developing embryos to environmental stress, these techniques are considered for the increased link between ARTs and imprinting disorders (Salvaing et al. 2016). It has been shown that there are differences in DNA methylation and transcript levels of number of genes in cord blood and placenta between children conceived using ART and children conceived in vivo (Song et al. 2015), an increased risk of some normally very rare imprinting disorders (Gosden et al. 2003) and epigenetic alterations at imprinted loci occur during ARTs (reviewed in (Hiura et al. 2012)). DNA methylation on cytosines (5-methylcytosine) is one of the vital epigenetic modification that can be affected from in vitro culture (Wright et al. 2011) and then may have an impact on the health of children born after ART.
DNA methylation mechanisms, de novo and maintenance methylation, are catalyzed by DNA methyltransferases (DNMTs) enzymes. Among structurally and functionally defined six different DNMTs, DNMT1 is primarily responsible from maintenance methylation and also contributes to de novo methylation process (Fatemi et al. 2002), and DNMT3A and DNMT3B are responsible for de novo methylation (Turek-Plewa and Jagodzinski 2005). It is known that DNA methylation has an essential role for development, growth and maturation of oocytes and early embryonic development during oogenesis and embryogenesis (Uysal et al. 2015). DNA methylation increases during oocyte maturation and then decrease after embryonic genome activation (Saitou et al. 2012).
Embryo culture media, which are considered as one of the most important environmental conditions that may affect embryo development, have been designed to mimic in vivo conditions under the guideline of many quality control tests to meet the standards expected by regulatory agencies and customers. Among them, mouse embryo assay (MEA), rises as a key tool to ensure quality of media as is functional and toxicological bioassay utilized to detect toxicity and suboptimal compounds (Esfandiari and Gubista 2020). Basically, MEA is set up to assess embryo morphology similarly to that used to score embryos in human ART. Besides quality control tests, media compositions are another issue that have been argued. Today, there are many commercially available embryo culture media, about which the question “whether any one medium or media system is superior to any other” raised (Sfontouris et al. 2016). Another popular question is which culture approach is better to increase ART success rates, the “back to nature” sequential approach, which was designed to supply identified changing metabolic and nutritional requirements of the developing embryo from zygote to blastocyst stage, or the “let the embryo choose” single medium approach, which let the developing embryos to choose required nutrients (Sfontouris et al. 2016). Generally, first step of sequential medium contains nonessential amino acids (NEAA) but not essentials (EAA) and high amount of pyruvate and lactate but low amount of glucose, for energy source (Morbeck et al. 2014), whereas single step media contains all amino acids and energy source molecules (Morbeck et al. 2017). Although, many studies aiming the answer those questions have been published in systematic review and meta-analysis of randomized controlled trials (Sfontouris et al. 2016), generally accepted idea is that there is insufficient evidence to recommend either sequential or single-step media as being superior for the culture of embryos to days 5/6, and future studies comparing these two media systems in well-designed trials should be performed.
In ART applications, retrieved oocytes can be seen in one of two maturation stages: mature metaphase II (MII) or immature either metaphase I (MI) or germinal vesicle (GV) oocytes. Immature oocytes are considered as the potential sources of embryos particularly when all or most of the retrieved oocytes are immature, and therefore those ones are used for intracytoplasmic sperm injection (ICSI). Immature oocytes were cultivated in embryo culture media after either in vitro maturation (IVM) of the oocytes in culture media or after immediate sperm injection, and several studies confirmed that both normal pregnancies and live births have been achieved from embryos originating from immature oocytes (Alvarez et al. 2013; Edirisinghe et al. 1997; Liu et al. 2003; Nagy et al. 1996; Shu et al. 2007).
In the current study, we aimed to evaluate the effects of two different embryo culture approach, sequential and single step culture media, on DNA methylating enzymes and global DNA methylation during embryo development and oocyte maturation. The fact that embryo culture media are designed for human IVF, they must pass the MEA test, before their release. Since there are many ethical and legal issues about human embryo experiments, and culture media supply optimal conditions for mouse embryo development, we implemented mouse model for this study. For embryo development studies, obtained zygotes were cultured in either sequential or single step culture media for 96 h, developed blastocysts were compared to in vivo developed ones obtained from super-ovulated mice. For oocyte maturation, retrieved GV stage oocytes were cultured either in first step media of sequential culture (FS-SM) approach or single step media (SSM) for 14–16 h. In vivo matured MII oocytes obtained from super-ovulated mice, and in vivo matured MII oocytes after mating with vasectomized males were also implemented as control and to evaluate the effect of super-ovulation. Distributions and amount of Dnmt1 and Dnmt3a enzymes were analyzed with immunofluorescence staining and Western blot analysis and global DNA methylation was evaluated with 5-mC staining for both embryo and oocyte experiments. Briefly, we found that signal intensity of Dnmt1 and Dnmt3a enzymes were high in single step media and control groups compared to sequential media during oocyte maturation and embryo development. These finding were supported with Western blot analysis. Similarly, global DNA methylation level in single step media group was comparable with the control but higher than the sequential media.
Materials and methods
Animals
This study was approved by the Animal Care and Usage Committee of Ankara University (protocol no: 2020-2-19). The female Balb/C mice (n = 60) at 4-weeks and male mice (n = 5) at 8–10 weeks of age were purchased from Research Animal Laboratory Unit of Ankara University School of Medicine. All mice were hosted with free access to food and water and kept in a 12 h light/dark cycle.
Collection and in vitro culture of zygotes and blastocyst collection
Detailed study design and media information was presented in Fig. 1 (Fig. 1). Briefly, the female mice were injected intraperitoneal (i.p.) with 5 IU pregnant mare’s serum gonadotropin (PMSG, Intervet, UK). Following 48 h of PMSG treatment, 5 IU human chorionic gonadotropin (hCG, Intervet, UK) were injected to the PMSG-primed female mice immediately mated overnight with mature male mice at a rate of 1 female: 1 male. The next morning, we checked the presence of vaginal plug. For control (in vivo group) blastocysts were obtained from uterus after 96 h of hCG injection of vaginal plug positive animals. For in vitro culture groups, zygotes were collected from oviducts after hCG injection at 20 h. Notably, the cumulus cells surrounding the 1-cell embryos were removed by using 1 mg/mL hyaluronidase (Sigma-Aldrich, USA) dissolved in GMOPS (Cat No: 10130, Vitrolife) medium. Zygotes were cultured in GTL as single step culture media (Cat No: 10134, Vitrolife) for Single step group; or in G1 (Cat No: 10128, Vitrolife, Sweden, first step of sequential media, FS-SM) and G2 (Cat No: 10132, Vitrolife, second step of sequential media, SS-SM) media as sequential culture group. For in vitro culture, embryos were cultured in 50 μl volumes of culture drops covered by OVOIL (Cat No: 10029, Vitrolife). Embryos at 4-cell stage in G1 media was transferred to G2 media, whereas no media replacement was applied for GTL group. All zygotes (0 h) were cultured to reach blastocyst stage (96 h) at 37 °C in an incubator with appropriate CO2 concentration providing media pH 7.2–7.4.
Study Design. Embryo experiments. After hormonal stimulation, female mice were mated with males. Vaginal plug positivity was considered as time zero. a In vivo: Blastocysts were collected from uterus (in vivo group as control). b Single: Obtained zygotes were cultured in GTL* for 96 h. Blastocyts were used for further experiments (Single step media group). c Seq: Obtained zygotes were cultured in G1* for 60 h (4 cell stage) and then in G2* for 36 h. Blastocyts were used for further experiments (Sequential media group). Oocyte experiments. a In Vivo/Vas: MII stage oocytes were collected from females mated with vasectomized males (in vivo group as control). b In Vivo/HS: MII stage oocytes were collected from hormonally stimulated females (in vivo group as hormone effect group). c Single: Obtained GV stage oocytes were in vitro cultured in GTL (Single step media group). d Seq: Obtained GV stage oocytes were in vitro cultured in G1 (First step of sequential media group). *G1 contains NEAA but not EAA, G2 NEAA and high EAA, GTL both NEAA and EAA (Morbeck et al. 2014, 2017)
Collection of oocytes and in vitro culture for oocyte maturation
Detailed study design was presented in Fig. 1 (Fig. 1). We designed two in vivo maturation groups. In the first one (In vivo/Vas group), MII oocytes were obtained from non-hormonally stimulated female mice after mating with vasectomized male mice. To evaluate the effect hormone stimulation in in vivo conditions (In Vivo/HS group), female mice were treated with 5 IU PMSG (i.p.) and subsequently i.p. injection of 5 IU hCG was administered 48 h after PMSG injection. We collected the cumulus-oocyte complexes (COCs) from oviducts’ ampulla region of the mice 14 h after hCG injection. The COCs were treated with 1 mg/mL hyaluronidase dissolved in GMOPS medium to remove cumulus cells, and the MII oocytes in normal morphology and having visible polar bodies are collected. Comparison of these two groups let us to analyze possible effect of superovulation. For in vitro groups, briefly, germinal vesicle (GV)-stage oocytes were collected from the ovaries of 4-week-old Balb/C female mice. For this purpose, after removing adipose tissue surrounding the ovaries, cumulus enclosed oocytes complexes (COC) containing fully grown GV-stage oocytes encircled by cumulus cells were released by puncturing the ovaries with a 23-gauge needle (Al-Zubaidi et al. 2021) in GMOPS medium. Attached cumulus cells were removed by repetitive pipetting of COCs. Denuded GV oocytes were cultured in GTL medium (single step group) or G1 medium as the first step of sequential culture condition (Seq group) covered by OVOIL at 37 °C with appropriate CO2 concentration for pH adjustment for 14–16 h.
Immunofluorescence (IF) staining
Blastocysts (n = 52 for Dnmt1, n = 57 for Dnmt3a, n = 53 for 5mC) and oocytes (n = 98 for Dnmt1, n = 92 for Dnmt3a, n = 65 for 5mC) were fixed in 4% paraformaldehyde (Sigma-Aldrich, USA) for 20 min, and then permeabilized with 1% Tween-20 (v/v) (Sigma-Aldrich, USA) prepared in phosphate buffered saline (PBS) for 15 min at room temperature. Just for 5mC (Cell Signaling, USA; 28692S) following permeabilization, the cells treated in 2 mol/L HCl for 30 min at RT (Liang et al. 2014). The cells in all groups were blocked in PBS containing 20% (v/v) normal goat serum (Vector Laboratory, USA) and then incubated in primary antibodies specific for either Dnmt1 (Abcam, USA; ab87654, reactive with either oocyte-specific (Dnmt1o) or somatic (Dnmt1s) isoforms), Dnmt3a (Abcam, USA; ab188470) or 5mC during overnight at 4 °C. After washing three times for 10 min with PBS including 2% (w/v) bovine serum albumin (BSA) (Sigma-Aldrich, USA), the cells were incubated with Alexa-488 conjugated anti-rabbit antibody (Invitrogen, USA) for 1 h at room temperature. After washing three times for 10 min with PBS-BSA, oocytes or embryos were gently transferred onto glass-bottomed 35-mm Petri dishes in a 4-μL drop of PBS-based mounting medium containing 1 μg/mL Hoechst 33,342 (Invitrogen, USA) for DNA labelling. The top was covered with paraffin oil. For negative control, PBS instead of primary antibody was used. All fluorescently tagged specimens were examined and imaged using a Zeiss LSM-880 Airyscan system (Zeiss, Germany) with 40 × Zeiss C-Apo water immersion objective (1.2 NA). Alexa-488 was excited using the 488 nm laser and a band pass of green-fluorescent emission was 493–634 nm. Hoechst 33,342 was excited with 405 nm laser and emission was collected with a band pass of 438–458 nm. Images were captured with Zeiss ZEN Black software. All laser, pinhole and gain parameters which were set based on the negative control were kept similar for each experiment.
Western blotting (WB)
Semi-quantitative analysis of Dnmt1 and Dnmt3a was performed with Western Blotting (WB) technique. For each group, embryos (n = 200) or oocytes (n = 250) were placed in lysis buffer (1% sodium dodecyl sulphate, 1 mmol/L sodium ortho-vanadate, 10 mmol/L Tris pH 7.4) supplemented with 1 × protease inhibitor cocktail (Amresco, USA). The protein concentration was measured using the BCA (bicinchoninic acid) method. Fifty micrograms of protein from each group was loaded on each lane of 10%Tris–HCl gel which was used for protein electrophoresis (BioRad, USA). Following electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Roche, UK) overnight at + 4 °C, the membrane was blocked with 5% (w/v) BSA prepared in TBS-T (20 mmol/L Tris/HCl and 150 m mol/L NaCl plus 0.05% Tween-20 at pH 7.4) at RT for 1 h. Membranes were incubated with primary antibodies specific to Dnmt1, Dnmt3a or β-actin (Abcam, USA) (1:1000 in 5% (w/v) BSA containing TBS-T) for 2 h at RT. Following a triple-wash in TBS-T for 15 min each, membranes were incubated with IRDye 800CW Goat anti-Rabbit IgG Secondary Antibody (1:2000 in TBS-T) (Licor Biosciences, USA) at RT for 1 h on a shaker. Protein band intensities were measured using a Li-Cor Odyssey CLx infrared detection system (LICOR Biosciences) following the manufacturer’s instructions.
Ratiometric image analysis
Ratiometric analysis was performed using ImageJ software (v.3.91, National Institutes of Health, Bethesda, Maryland, USA). Briefly, Dnmt (green) and DNA (blue) channels of confocal images were separated, and the green one, which were later used for signal measurement, converted to 32-bit images. Images obtained from Li-Cor Odyssey CLx infrared detection system were converted to 32-bit images. Total signal intensities as gray value from each pixel were measured from all oocytes and embryos for microscopic images, and from region of interest for WB images using the Image Calculation function in ImageJ software (Al-Zubaidi et al. 2021; Uysal et al. 2021). The background signals were used for thresholding.
Statistical analysis
All experiments were repeated at least in three times. One-way analysis of variance (ANOVA) followed by Dunn’s post hoc test was performed by using SigmaStat for Windows, version 3.5 (Jandel Scientific Corp). For all tests, p < 0.05 was considered to be statistically significant.
Results
Effects of culture media on Dnmts and DNA methylation during embryo development
A diffuse Dnmt1 staining was observed at the cytoplasm (Fig. 2a). Dnmt3a localized at the cytoplasm of embryoblasts, whereas at the cytoplasm and nucleus of trophoblast (Fig. 3a). 5mC signal was detected at the nuclei of all cells (Fig. 4a). We did not observe any difference in the staining pattern in both Dnmt1 and Dnmt3a, and 5mC in all groups.
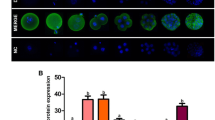
Representative images for immunofluorescent (IF) staining (a) and Western blotting (WB) (c). Dnmt1 (green) and DNA (blue, in IF) for blastocyst stage embryos obtained from IV: In vivo developed embryos group, Single: Single step medium, Seq: Sequential medium. Mean ± S.D for the total signal intensity from each pixel of Dnmt1 from IF (b) and relative signal intensity of Dnmt1 to actin from WB (d). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 20 µm
Demonstrative images for immunofluorescent (IF) staining (a) and Western blotting (WB) (c). Dnmt3a (green) and DNA (blue, in IF) for blastocyst stage embryos obtained from IV In vivo developed embryos group, Single Single step medium, Seq Sequential medium. Mean ± S.D for the total signal intensity from each pixel of Dnmt3a from IF (b) and relative signal intensity of Dnmt3a to actin from WB (d). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 20 µm
Representative images for immunofluorescent (IF) staining of 5mC (green) staining (a) for blastocyst stage embryos obtained from IV In vivo developed embryos group, Single Single step medium, Seq Sequential medium. Mean ± S.D for the total signal intensity from each pixel of 5mC (b). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 20 µm
Analyses of total fluorescence signal intensities of Dnmt1 signals demonstrated that control (in vivo group) and Single groups were comparable but Seq group was significantly low (p < 0.05) (Fig. 2b). In parallel with IF outcomes, WB analysis (Fig. 2c) revealed that Dnmt1 levels were similar in control and Single groups, but a significant decrease exists in Seq group (p < 0.05) (Fig. 2d).
We observed similar outcomes from Dnmt3a to Dnmt1 as total fluorescence signal intensities of Dnmt3a were comparable for control and Single group but significantly low in Seq group (Fig. 3b). WB analysis supported this finding (Fig. 3c and d).
When global DNA methylation was assessed with 5mC staining, we observed that decreased staining in Seq group (p < 0.05), whereas comparable results for control and Single groups (Fig. 4b).
Effects of culture media on Dnmts and global DNA methylation during oocyte maturation
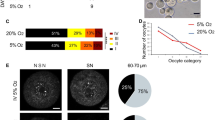
Strong Dnmt1 signals were found just beneath the oolemma while remaining cytoplasm found lack of the staining (Fig. 5a), whereas Dnmt3a staining was diffuse and also punctate in some loci of cytoplasm (Fig. 6a). 5mC staining was seen at the chromosomes located in MII spindle (Fig. 7a). Similar distribution and localization pattern of Dnmt1 and 3a, and 5mC were noted in all groups.
Representative images for immunofluorescent (IF) staining (a) and Western blotting (WB) (c). Dnmt1 (green) and DNA (blue, in IF) for MII stage oocytes obtained from In vivo/Vas: In vivo matured MII stage oocytes obtained from female mice mated with vasectomised males, In Vivo/HS In vivo matured MII stage oocytes obtained from Hormonally Stimulated female mice, Single Single step medium, Seq Sequential medium. Mean ± S.D for the total signal intensity from each pixel of Dnmt1 from IF (b) and relative signal intensity of Dnmt1 to actin from WB (d). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 10 µm
Demonstrative images for immunofluorescent (IF) staining (a) and Western blotting (WB) (c). Dnmt3a (green) and DNA (blue, in IF) for MII stage oocytes obtained from In vivo/Vas: In vivo matured MII stage oocytes obtained from female mice mated with vasectomised males, In Vivo/HS In vivo matured MII stage oocytes obtained from Hormonally Stimulated female mice, Single Single step medium, Seq Sequential medium. Mean ± S.D for the total signal intensity from each pixel of Dnmt3a from IF (b) and relative signal intensity of Dnmt3a to actin from WB (d). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 10 µm
Representative images for immunofluorescent (IF) staining of 5mC (green) staining (a) for MII stage oocytes obtained from In vivo/Vas In vivo matured MII stage oocytes obtained from female mice mated with vasectomised males, In Vivo/HS In vivo matured MII stage oocytes obtained from Hormonally Stimulated female mice, Single Single step medium, Seq Sequential medium. Mean ± S.D for the total signal intensity from each pixel of 5mC (b). Different letters indicate p < 0.05. DIC Differential Interference Contrast, NC Negative control. Scale bar: 10 µm
Total fluorescence signal intensity analysis of Dnmt1 signals revealed that control (In Vivo/Vas group), In Vivo/HS and Single groups were comparable but Seq group was significantly lower than the others (p < 0.05) (Fig. 5b). Similarly, WB analysis (Fig. 5c) showed that Dnmt1 levels were not different between In Vivo/Vas, In Vivo/HS and Single groups, but a significant decrease was noted in Seq group (p < 0.05) (Fig. 5d).
We found similar results for Dnmt3a as total fluorescence signal intensities of Dnmt3a were comparable for In Vivo/Vas, In Vivo/HS and Single groups but significantly low in Seq group (Fig. 6b). Similar pattern was noted from WB analysis (Fig. 6c and d).
Total fluorescence signal intensity of 5mC was comparable in In Vivo/Vas, In Vivo/HS and Single groups, but Seq group was significantly (p < 0.05) lower than the others (Fig. 7a and b).
Discussion
DNA methylation, one of the major epigenetic modification of the genome, occurs by the activities of specific enzymes, DNA methyltransferases (DNMTs), which can add methyl groups to the fifth carbon atom of the cytosine residues within cytosine-phosphate guanine (CpG) and non-CpG dinucleotides sites (Reik & Dean 2001). Besides many cells and tissues, this epigenetic modification plays vital roles in the control of gene expression during oogenesis and early embryogenesis (Saitou et al. 2012).
In the current study, we demonstrated that the culture media which are routinely utilized in ART laboratories for embryo development and/or oocyte maturation under the certain conditions, affect the expression levels of Dnmt1 and Dnmt3a, and also global DNA methylation in oocytes and pre-implantation stage mouse embryos. Levels of Dnmts enzymes and global DNA methylation in single step media was comparable with in vivo matured oocytes obtained from female mice after either superovulation or mating with the vasectomized males, whereas significantly low in the first step of sequential media compared to other groups. Similarly, Dnmts and global DNA methylation levels of in vivo cultured embryos in single step media was comparable with the in vivo developed embryos but significantly low in in vivo cultured embryos in sequential media. Furthermore, for embryo development, decrease in Dnmt1 and Dnmt3a levels and also 5mC with first step medium, was not compensated with second step medium even though its’ composition is similar to single step medium.
We think that glucose may be one of the factors affecting Dnmts and global DNA methylation since concentrations of glucose show significant variations between the culture media as 0.97 mM in Single step, 0.5 in FS-SM and 3.4 in SS-SM. It was shown that high-glucose concentrations (10 mM and 15 mM) did not alter DNA methylation levels of H19, whereas significantly increased the DNA methylation levels of the maternally imprinted gene Peg3 and decreased the levels in the promoter of adiponectin in human IVM (Wang et al. 2018). Recently, Li et al. (Li et al. 2020) analysed the effect of type 1 diabetes on zona pellucida and genomic DNA methylation of oocytes and granulosa cells in streptozocin induced mouse model and found that Hmg20b, Id4, Pi16, Rab3a, Ramp1, and Shank3 genes in oocytes were differentially methylated, and differentially hypermethylated genes and differentially hypomethylated genes in oocytes involved 13 areas including biological adhesion, cell proliferation, developmental processes, metabolic processes, and reproduction. On the other hand, it was not found any differences in the expression of mRNA encoding for DNMT3 between in vivo and in vitro developed equine embryos treated with 5–10 mM of glucose during culture, while apoptosis, mitochondrial function, and glucose metabolism genes were affected (Sanchez-Calabuig et al. 2021). Tremblay et al. (Tremblay et al. 2018) generated zygotes from cow oocytes and then exposed to high (5 mM) glucose concentration whereas 0.2 mM in control conditions for the first 3 days post-fertilization and then followed by normal media until the blastocyst stage. They observed that elevated glucose led to hypomethylation close to telomeric regions and methylation changes on genomic regions associated with energy metabolism.
Bhagat and et al. (Bhagat et al. 2019) demonstrated that lactate produced by neoplastic cells leads to increased production of alpha-ketoglutarate which mediated activation of the demethylase TET enzyme led to decreased cytosine methylation and increased hydroxymethylation during de novo differentiation of mesenchymal stem cells to cancer associated fibroblasts, whereas Thangaraju et al. (Thangaraju et al. 2006) showed inhibitory effect of pyruvate, but not lactate, on histone deacetylases in MCF7 breast tumor cells. These studies present the possible effects of lactate and pyruvate, which are vital energy sources for oocyte maturation and pre-zygotic genome activation stage embryos, on epigenetic programming via different mechanism. Therefore, we think that lactate/pyruvate ratio, which shows significant differences between the media as 18 in SSM, 36 in FS-SM and 86 in SS-SM, may be another factor that may affect Dnmt levels and also global DNA methylation, however, we did not find any study in the literature which focus on the possible effect of lactate/pyruvate ratio on DNA methylation of oocyte and embryo to support or reject our hypothesis.
Amino acid compositions of embryo culture media show significant differences, as Single step medium contains both EAA and NEAA, whereas first step medium consists of NEAA not EAA and second step medium includes both EAA and NEAA with the different concentration from the single step (Morbeck et al. 2014, 2017). Park et al. (Park et al. 2014) analyzed the effects of EAA and/or NEAA on certain imprinting genes in parthenogenetically activated porcine embryos and found that the expression of H19 increased while expression of IGF2R decreased in response to EAA, whereas NNAT expression decreased in response to NEAA. Additionally, they noted that EAA increased the expression of PEG1, and this enhanced expression can be compensated by the inclusion of NEAA.
Salvaing et al. (Salvaing et al. 2016) analyzed the levels of methylation and hydroxymethylation, by immunostaining, and also expression of Dnmt1, 3a, and 3b enzymes, by qPCR in rabbit embryos which in vitro cultured either in one step medium or sequential media obtained from different media producers, and then compared their results with in vivo developed embryos. They did not find significant differences in the expression of Dnmt1 and 3a levels in embryos between the groups. On the other hand, we found significant differences between sequential media and single step and in vivo group by immunostaining and western blot at blastocyst stage embryos. We think that these discrepancies between two studies can be caused by either the utilized techniques, which qPCR vs. immunostaining and western blot, or species dependent, as rabbit vs. mouse. They also noted significant differences in DNA methylation between the groups in a stage dependent manner until the morula stage, while blastocyst comparison was not presented in the manuscript. On the other hand, in the current study, along with the oocyte maturation, we aimed to focus blastocyst stage embryos which are prone to implant to the uterus after hatching.
Saenz-de-Juano et al. (Saenz-de-Juano et al. 2019) presented genome-wide analysis of DNA methylation in MII oocytes obtained after natural ovulation, in vitro follicle culture and superovulation, and concluded that in vitro follicle culture is associated with altered methylation at specific set of loci. They also added that DNA methylation of superovulated oocytes obtained from prepubertal mice differs from the that of adult oocytes, while oocytes from superovulated adult females or naturally ovulated oocytes show slight differences. Similarly, Huo et al. (Huo et al. 2020) analyzed the effect of different dosages of FSH/hMG combined with hCG in female mice and found that superovulation causes methylation alterations at a specific set of loci, and the differentially methylated regions mainly occurred in regions other than promoters many of which are involved in vital biological processes such as glucose metabolism, nervous system development, cell cycle, cell proliferation, and embryo implantation. In the current study, we did not find any differences between IV group, in which mice were hormonally (5 IU PMSG and HCG) stimulated, and Vas group, not hormonally stimulated, in Dnmt1 and 3a and also global DNA methylation in MII stage oocytes. Our findings supported Uysal et al.’s study (Uysal et al. 2018) in which researchers did not find significant difference in Dnmt1, 3a and 3b levels between control and 5 IU PMSG and HCG stimulated groups whereas, they found that high dose hormone application (7.5 IU PMSG and HCG) caused significant decrease in some of those enzyme levels in oocytes and early embryos in a stage dependent manner. Furthermore, in the current study, we did not observe significant difference in global DNA methylation between control and hormonally stimulated groups.
In conclusion, this is the first study evaluating the effects of embryo culture media on DNA methylating enzymes and global DNA methylation on in vitro developed embryos and in vitro matured oocytes. Due to ethical and legal concerns, using human embryos or oocytes would not be possible to demonstrate effect of culture media on DNA methylation during in vitro culture. As culture media utilized in human ART require MEA test to appraise if it is appropriate for clinical usage, we think that mouse model may be a good option for our study design. In this study, that we utilize media from one single producer is important to get rid of the concern any possible media supplier effect on the study. We think that media from other producers and human oocytes/embryo experiments still needs to be done to clarify the issue, and necessity for future studies raised to assess whether these alterations in Dnmts and global DNA methylation are related to specific methylation changes and can be physiologically significant or not.
Data availability
Not applicable.
Code availability
Not applicable.
References
Alvarez C, Garcia-Garrido C, Taronger R, Gonzalez de Merlo G (2013) In vitro maturation, fertilization, embryo development & clinical outcome of human metaphase-I oocytes retrieved from stimulated intracytoplasmic sperm injection cycles. Indian J Med Res 137(2):331–338
Al-Zubaidi U, Adhikari D, Cinar O, Zhang QH, Yuen WS, Murphy MP, Rombauts L, Robker RL, Carroll J (2021) Mitochondria-targeted therapeutics, MitoQ and BGP-15, reverse aging-associated meiotic spindle defects in mouse and human oocytes. Hum Reprod 36(3):771–784. https://doi.org/10.1093/humrep/deaa300
Bhagat TD, Von Ahrens D, Dawlaty M, Zou Y, Baddour J, Achreja A, Zhao H, Yang L, Patel B, Kwak C, Choudhary GS, Gordon-Mitchell S, Aluri S, Bhattacharyya S, Sahu S, Bhagat P, Yu Y, Bartenstein M, Giricz O, Suzuki M, Sohal D, Gupta S, Guerrero PA, Batra S, Goggins M, Steidl U, Greally J, Agarwal B, Pradhan K, Banerjee D, Nagrath D, Maitra A, Verma A (2019) Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. Elife. https://doi.org/10.7554/eLife.50663
Edirisinghe WR, Junk SM, Matson PL, Yovich JL (1997) Birth from cryopreserved embryos following in-vitro maturation of oocytes and intracytoplasmic sperm injection. Hum Reprod 12(5):1056–1058. https://doi.org/10.1093/humrep/12.5.1056
Esfandiari N, Gubista A (2020) Mouse embryo assay for human in vitro fertilization quality control: a fresh look. J Assist Reprod Genet 37(5):1123–1127. https://doi.org/10.1007/s10815-020-01768-9
Fatemi M, Hermann A, Gowher H, Jeltsch A (2002) Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem 269(20):4981–4984. https://doi.org/10.1046/j.1432-1033.2002.03198.x
Gosden R, Trasler J, Lucifero D, Faddy M (2003) Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet 361(9373):1975–1977. https://doi.org/10.1016/S0140-6736(03)13592-1
Hiura H, Okae H, Miyauchi N, Sato F, Sato A, Van De Pette M, John RM, Kagami M, Nakai K, Soejima H, Ogata T, Arima T (2012) Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproduction technologies. Hum Reprod 27(8):2541–2548. https://doi.org/10.1093/humrep/des197
Huo Y, Yan ZQ, Yuan P, Qin M, Kuo Y, Li R, Yan LY, Feng HL, Qiao J (2020) Single-cell DNA methylation sequencing reveals epigenetic alterations in mouse oocytes superovulated with different dosages of gonadotropins. Clin Epigenetics 12(1):75. https://doi.org/10.1186/s13148-020-00866-w
Li L, Jing Y, Dong MZ, Fan LH, Li QN, Wang ZB, Hou Y, Schatten H, Zhang CL, Sun QY (2020) Type 1 diabetes affects zona pellucida and genome methylation in oocytes and granulosa cells. Mol Cell Endocrinol. https://doi.org/10.1016/j.mce.2019.110627
Liang Y, Fu XW, Li JJ, Yuan DS, Zhu SE (2014) DNA methylation pattern in mouse oocytes and their in vitro fertilized early embryos: effect of oocyte vitrification. Zygote 22(2):138–145. https://doi.org/10.1017/S0967199412000512
Liu J, Lu G, Qian Y, Mao Y, Ding W (2003) Pregnancies and births achieved from in vitro matured oocytes retrieved from poor responders undergoing stimulation in in vitro fertilization cycles. Fertil Steril 80(2):447–449. https://doi.org/10.1016/s0015-0282(03)00665-4
Morbeck DE, Krisher RL, Herrick JR, Baumann NA, Matern D, Moyer T (2014) Composition of commercial media used for human embryo culture. Fertil Steril. https://doi.org/10.1016/j.fertnstert.2014.05.043
Morbeck DE, Baumann NA, Oglesbee D (2017) Composition of single-step media used for human embryo culture. Fertil Steril. https://doi.org/10.1016/j.fertnstert.2017.01.007
Nagy ZP, Cecile J, Liu J, Loccufier A, Devroey P, Van Steirteghem A (1996) Pregnancy and birth after intracytoplasmic sperm injection of in vitro matured germinal-vesicle stage oocytes: case report. Fertil Steril 65(5):1047–1050. https://doi.org/10.1016/s0015-0282(16)58285-5
Park CH, Jeong YH, Jeong YI, Kwon JW, Shin T, Hyun SH, Jeung EB, Kim NH, Seo SK, Lee CK, Hwang WS (2014) Amino acid supplementation affects imprinted gene transcription patterns in parthenogenetic porcine blastocysts. PLoS One 9(9):e106549. https://doi.org/10.1371/journal.pone.0106549
Reik W, Dean W (2001) DNA methylation and mammalian epigenetics. Electrophoresis 22(14):2838–2843. https://doi.org/10.1002/1522-2683(200108)22:14%3c2838::AID-ELPS2838%3e3.0.CO;2-M
Saenz-de-Juano MD, Ivanova E, Billooye K, Herta AC, Smitz J, Kelsey G, Anckaert E (2019) Genome-wide assessment of DNA methylation in mouse oocytes reveals effects associated with in vitro growth, superovulation, and sexual maturity. Clin Epigenetics 11(1):197. https://doi.org/10.1186/s13148-019-0794-y
Saitou M, Kagiwada S, Kurimoto K (2012) Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139(1):15–31. https://doi.org/10.1242/dev.050849
Salvaing J, Peynot N, Bedhane MN, Veniel S, Pellier E, Boulesteix C, Beaujean N, Daniel N, Duranthon V (2016) Assessment of “one-step” versus “sequential” embryo culture conditions through embryonic genome methylation and hydroxymethylation changes. Hum Reprod 31(11):2471–2483. https://doi.org/10.1093/humrep/dew214
Sanchez-Calabuig MJ, Fernandez-Gonzalez R, Hamdi M, Smits K, Lopez-Cardona AP, Serres C, Macias-Garcia B, Gutierrez-Adan A (2021) A high glucose concentration during early stages of in vitro equine embryo development alters expression of genes involved in glucose metabolism. Equine Vet J 53(4):787–795. https://doi.org/10.1111/evj.13342
Sfontouris IA, Martins WP, Nastri CO, Viana IG, Navarro PA, Raine-Fenning N, van der Poel S, Rienzi L, Racowsky C (2016) Blastocyst culture using single versus sequential media in clinical IVF: a systematic review and meta-analysis of randomized controlled trials. J Assist Reprod Genet 33(10):1261–1272. https://doi.org/10.1007/s10815-016-0774-5
Shu Y, Gebhardt J, Watt J, Lyon J, Dasig D, Behr B (2007) Fertilization, embryo development, and clinical outcome of immature oocytes from stimulated intracytoplasmic sperm injection cycles. Fertil Steril 87(5):1022–1027. https://doi.org/10.1016/j.fertnstert.2006.08.110
Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, Coutifaris C, Sapienza C (2015) DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics 7:41. https://doi.org/10.1186/s13148-015-0071-7
Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, Sterneck E, Ganapathy V (2006) SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res 66(24):11560–11564. https://doi.org/10.1158/0008-5472.CAN-06-1950
Tremblay R, Dufort I, Sirard MA (2018) Metabolic stress induces modifications in the epigenetic program of preimplantation bovine embryos. Mol Reprod Dev 85(2):117–127. https://doi.org/10.1002/mrd.22941
Turek-Plewa J, Jagodzinski PP (2005) The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 10(4):631–647
Uysal F, Akkoyunlu G, Ozturk S (2015) Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos. Biochimie 116:103–113. https://doi.org/10.1016/j.biochi.2015.06.019
Uysal F, Ozturk S, Akkoyunlu G (2018) Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J Assist Reprod Genet 35(3):503–513. https://doi.org/10.1007/s10815-017-1087-z
Uysal F, Cinar O, Can A (2021) Knockdown of Dnmt1 and Dnmt3a gene expression disrupts preimplantation embryo development through global DNA methylation. J Assist Reprod Genet. https://doi.org/10.1007/s10815-021-02316-9
Wang Q, Tang SB, Song XB, Deng TF, Zhang TT, Yin S, Luo SM, Shen W, Zhang CL, Ge ZJ (2018) High-glucose concentrations change DNA methylation levels in human IVM oocytes. Hum Reprod 33(3):474–481. https://doi.org/10.1093/humrep/dey006
Wright K, Brown L, Brown G, Casson P, Brown S (2011) Microarray assessment of methylation in individual mouse blastocyst stage embryos shows that in vitro culture may have widespread genomic effects. Hum Reprod 26(9):2576–2585. https://doi.org/10.1093/humrep/der201
Acknowledgements
This work was supported by The Scientific and Technological Research Council of Turkey (Grant number 120S175). Preliminary outcomes of the study were submitted to present in 13th Biennial Alpha Conference, 2020, Sevilla, Spain. Although it was accepted, it has not been presented yet, as the meeting postponed due to Covid-19 pandemic.
Funding
This work was supported by The Scientific and Technological Research Council of Turkey (Grant Number 120S175).
Author information
Authors and Affiliations
Contributions
All authors qualify for authorship by contributing substantially to this article. FU and OC developed the original concept of this study and wrote the manuscript. All authors collected data and FU performed statistical analysis. All authors have contributed to critical discussion, reviewed the final version of the article, and approved it for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
The experimental protocol was approved by the Animal Care and Usage Committee of Ankara University (protocol no: 2020-2-19).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Uysal, F., Kahveci, S., Sukur, G. et al. Embryo culture media differentially alter DNA methylating enzymes and global DNA methylation in embryos and oocytes. J Mol Histol 53, 63–74 (2022). https://doi.org/10.1007/s10735-021-10038-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-021-10038-6