Abstract
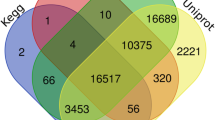
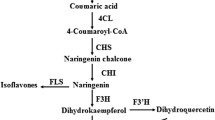
The inflorescence of Chrysanthemum morifolium cv. ‘Hangju’ has been widely used in China due to its antioxidant and anti-inflammatory properties. The biosynthesis and regulation of flavonoids, a group of bioactive components, in C. morifolium are poorly understood. Transcriptome sequencing is an effective method for obtaining transcript information. Therefore, single-molecule real-time (SMRT) sequencing was performed to obtain the full-length genes involved in flavonoid biosynthesis and regulation in C. morifolium. High-quality RNA was extracted from the inflorescence of C. morifolium at different flowering stages and used to construct two libraries (0–5 kb and 4.5–10 kb) for sequencing. Finally, 125,532 non-redundant isoforms with a mean length of 2009 bp were obtained. Of these, 2,083 transcripts were annotated to pathways related to flavonoid biosynthesis, and 56 isoforms were annotated as CHS, CHI, F3H, F3’H, FNS II, FLS, DFR and ANS genes. Based on gene expression levels at different stages, we predicted the major genes involved in flavonoid biosynthesis. By phylogenetic analysis, we found two candidate MYB transcription factors (CmMYBF1 and CmMYBF2) activating flavonol biosynthesis. Based on the full-length transcriptomic data and further quantitative analysis, the major genes involved in flavonoid biosynthesis and regulation in C. morifolium were predicted in our study. The results provide a valuable theoretical basis for the introduction and cultivation of C. morifolium cv. ‘Hangju’.




Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to the data was analyzing for another research, but are available from the corresponding author on reasonable request.
Abbreviations
- SMRT-Seq:
-
Single-molecule real-time sequencing
- EBGs:
-
Early biosynthetic genes
- LBGs:
-
Late biosynthetic genes
- SGS:
-
Second-generation sequencing
- TGS:
-
Third-generation sequencing
- ROI:
-
Reads of insert
- NR:
-
Non-redundant protein
- NT:
-
Non-redundant nucleotide
- GO:
-
Gene Ontology
- KOG:
-
Clusters of Eukaryotic Orthologous Groups
- KEGG:
-
Kyoto Encyclopaedia of Genes and Genomes
- ICE:
-
Interative Clustering and Error Correction
- PAL:
-
Phenylalanine ammonia-lyase
- C4H:
-
Cinnamate 4-hydroxylase
- 4CL:
-
4-Coumarate-CoA ligase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3H:
-
Flavanone 3-hydroxylase
- F3’H:
-
Flavonoid 3’-hydroxylase
- F3’5’H:
-
Flavonoid 3’,5’-hydroxylase
- FNS II:
-
Flavone synthase II
- IFS:
-
Isoflavone synthase
- FLS:
-
Flavonol synthase
- DFR:
-
Dihydroflavonol 4-reductase
- ANS:
-
Anthocyanidin synthase
- ANR:
-
Anthocyanidin reductase
- LAR:
-
Leucoanthocyanidin reductase
- MBW:
-
MYB-bHLH-WD40
- SG:
-
Subgroup
- ORF:
-
Open read frame
- AS:
-
Alternative splicing
- FL:
-
Full length
References
Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy AS (2016) A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun 7:11706. https://doi.org/10.1038/ncomms11706
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39(3):366–380. https://doi.org/10.1111/j.1365-313X.2004.02138.x
Cheng B, Furtado A, Henry RJ (2017) Long-read sequencing of the coffee bean transcriptome reveals the diversity of full-length transcripts. Gigascience 6(11):1–13. https://doi.org/10.1093/gigascience/gix086
Cui G, Niu Y, Wang H, Dong J, Yuki H, Chen S (2012) Rapid isolation and identification of active antioxidant ingredients from Gongju using HPLC-DAD-ESI-MS(n) and postcolumn derivatization. J Agric Food Chem 60(21):5407–5413. https://doi.org/10.1021/jf300938e
Deng YX, Lu SF (2017) Biosynthesis and regulation of phenylpropanoids in plants. Crit Rev Plant Sci 36(4):257–290. https://doi.org/10.1080/07352689.2017.1402852
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49(3):414–427. https://doi.org/10.1111/j.1365-313X.2006.02964.x
Gao T, Zhu ZY, Zhou X, Xie ML (2016) Chrysanthemum morifolium extract improves hypertension-induced cardiac hypertrophy in rats by reduction of blood pressure and inhibition of myocardial hypoxia inducible factor-1alpha expression. Pharm Biol 54(12):2895–2900. https://doi.org/10.1080/13880209.2016.1190764
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827. https://doi.org/10.1111/j.1365-313X.2007.03373.x
He D, Ru X, Wen L, Wen Y, Jiang H, Bruce IC, Jin J, Ma X, Xia Q (2012) Total flavonoids of Flos Chrysanthemi protect arterial endothelial cells against oxidative stress. J Ethnopharmacol 139(1):68–73. https://doi.org/10.1016/j.jep.2011.10.043
Hu T, Gao ZQ, Hou JM, Tian SK, Zhang ZX, Yang L, Liu Y (2020) Identification of biosynthetic pathways involved in flavonoid production in licorice by RNA-seq based transcriptome analysis. Plant Growth Regul 92(1):15–28. https://doi.org/10.1007/s10725-020-00616-1
Jin HF, Liu XW, Tang YM, Tang LJ, Wang YL, Du CQ (2016) Effects of total flavones from Dendranthema morifolium on vasocontraction and proliferation of vascular smooth muscle cells. Mol Med Rep 13(1):989–993. https://doi.org/10.3892/mmr.2015.4576
Jung W, Yu O, Lau SMC, O’Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18(2):208–212. https://doi.org/10.1038/72671
Kuang CL, Lv D, Shen GH, Li SS, Luo QY, Zhang ZQ (2018) Chemical composition and antimicrobial activities of volatile oil extracted from Chrysanthemum morifolium Ramat. J Food Sci Technol 55(7):2786–2794. https://doi.org/10.1007/s13197-018-3203-1
Liu H, Sun M, Du D, Pan H, Cheng T, Wang J, Zhang Q (2015a) Whole-transcriptome analysis of differentially expressed genes in the vegetative buds, floral buds and buds of Chrysanthemum morifolium. PLoS ONE 10(5):e0128009. https://doi.org/10.1371/journal.pone.0128009
Liu XF, Xiang LL, Yin XR, Grierson D, Li F, Chen KS (2015b) The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Sci Hortic 194:278–285. https://doi.org/10.1016/j.scienta.2015.08.018
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138(2):1083–1096. https://doi.org/10.1104/pp.104.058032
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteomics Bioinform 13(5):278–289. https://doi.org/10.1016/j.gpb.2015.08.002
Righini S, Rodriguez EJ, Berosich C, Grotewold E, Casati P, Ferreyra MLF (2019) Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant Cell Environ 42(2):495–508. https://doi.org/10.1111/pce.13428
Shao QS, Guo QS, Deng YM, Guo HP (2010) A comparative analysis of genetic diversity in medicinal Chrysanthemum morifolium based on morphology, ISSR and SRAP markers. Biochem Syst Ecol 38(6):1160–1169. https://doi.org/10.1016/j.bse.2010.11.002
Sharon D, Tilgner H, Grubert F, Snyder M (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31(11):1009–1014. https://doi.org/10.1038/nbt.2705
Song C, Liu Y, Song A, Dong G, Zhao H, Sun W, Ramakrishnan S, Wang Y, Wang S, Li T, Niu Y, Jiang J, Dong B, Xia Y, Chen S, Hu Z, Chen F, Chen S (2018) The Chrysanthemum nankingense genome provides insights into the evolution and diversification of Chrysanthemum flowers and medicinal traits. Mol Plant 11(12):1482–1491. https://doi.org/10.1016/j.molp.2018.10.003
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456. https://doi.org/10.1016/s1369-5266(00)00199-0
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50(4):660–677. https://doi.org/10.1111/j.1365-313X.2007.03078.x
Tao JH, Duan JA, Jiang S, Feng NN, Qiu WQ, Ling Y (2017) Polysaccharides from Chrysanthemum morifolium Ramat ameliorate colitis rats by modulating the intestinal microbiota community. Oncotarget 8(46):80790–80803. https://doi.org/10.18632/oncotarget.20477
Ukiya M, Akihisa T, Yasukawa K, Kasahara Y, Kimura Y, Koike K, Nikaido T, Takido M (2001) Constituents of compositae plants. 2. Triterpene diols, triols, and their 3-o-fatty acid esters from edible chrysanthemum flower extract and their anti-inflammatory effects. J Agric Food Chem 49(7):3187–3197. https://doi.org/10.1021/jf010164e
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3(1):2–20. https://doi.org/10.1093/mp/ssp106
Wang T, Zhu ZB, Guo QS, Mao PF (2013) Variation in major flavonoids glycosides and caffeoylquinic acids during florescence of three Chrysanthemum morifolium Ramat cv. ‘Hangju’ genotypes. Biochem Syst Ecol 47:74–79. https://doi.org/10.1016/j.bse.2012.11.004
Wang T, Guo QS, Mao PF (2014) Flavonoid accumulation during florescence in three Chrysanthemum morifolium Ramat cv. ‘Hangju’ genotypes. Biochem Syst Ecol 55:79–83. https://doi.org/10.1016/j.bse.2014.02.023
Wang K, Bai ZY, Liang QY, Liu QL, Zhang L, Pan YZ, Liu GL, Jiang BB, Zhang F, Jia Y (2018) Transcriptome analysis of chrysanthemum (Dendranthema grandiflorum) in response to low temperature stress. BMC Genomics 19(1):319. https://doi.org/10.1186/s12864-018-4706-x
Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1(3):251–257. https://doi.org/10.1016/s1369-5266(98)80113-1
Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, Chen KS (2015) A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 10(11):e0143892. https://doi.org/10.1371/journal.pone.0143892
Xu Y, Gao S, Yang Y, Huang M, Cheng L, Wei Q, Fei Z, Gao J, Hong B (2013) Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics 14:662. https://doi.org/10.1186/1471-2164-14-662
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20(3):176–185. https://doi.org/10.1016/j.tplants.2014.12.001
Xu PB, Zawora C, Li Y, Wu J, Liu LC, Liu ZC, Cai R, Lian HL (2018) Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul 86(1):121–132. doi:https://doi.org/10.1007/s10725-018-0415-3
Yue J, Zhu C, Zhou Y, Niu X, Miao M, Tang X, Chen F, Zhao W, Liu Y (2018) Transcriptome analysis of differentially expressed unigenes involved in flavonoid biosynthesis during flower development of Chrysanthemum morifolium ‘Chuju’. Sci Rep 8(1):13414. doi:https://doi.org/10.1038/s41598-018-31831-6
Zhao N, Li C, Yan Y, Cao W, Song A, Wang H, Chen S, Jiang J, Chen F (2018) Comparative Transcriptome analysis of waterlogging-sensitive and waterlogging-tolerant Chrysanthemum morifolium cultivars under waterlogging stress and reoxygenation conditions. Int J Mol Sci 19(5):21. https://doi.org/10.3390/ijms19051455
Zheng C, Dong Q, Chen H, Cong Q, Ding K (2015) Structural characterization of a polysaccharide from Chrysanthemum morifolium flowers and its antioxidant activity. Carbohydr Polym 130:113–121. https://doi.org/10.1016/j.carbpol.2015.05.004
Zhou LL, Shi MZ, Xie DY (2012) Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta 236(3):825–837. https://doi.org/10.1007/s00425-012-1674-2
Zou QJ, Wang T, Guo QS, Zhang WY, Wang R, Wang YH, Xiao YM (2018) [Prokaryotic expression and in vitro enzyme activity analysis of flavonol synthase in Chrysanthemum morifolium cv. ‘Hangju.’ China J Chin Mater Med 43(17):3471–3476. https://doi.org/10.19540/j.cnki.cjcmm.20180510.007
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National Nature Science Foundation of China (No. 81503180) in the design of the study and collection of data. The Fundamental Research Funds for the Central Universities (No. KYZ201608 and KJQN201643) were used in supporting the analysis and interpretation of data and manuscript writing.
Author information
Authors and Affiliations
Contributions
T.W., F.Y., and Q.G. conceived and designed the experiments. T.W. and F.Y. performed the experiments and written the original draft. T.W., F.Y., and Q.Z. analyzed the data and make figures and tables. W.Z. and L.Z. contributed to the cultivation and collection of experimental materials. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2020_660_MOESM5_ESM.pdf
Supplementary Fig. 1 The electrophoretogram of RNA isolated from inflorescence at five different flowering stages (PDF 346.8 kb)
10725_2020_660_MOESM8_ESM.pdf
Supplementary Fig. 4 The phylogenetic tree of MYB factors between C. morifolium and Arabidopsis thaliana. The MYB factors clustered into R2R3-MYB subgroup 4-7 regulating the flavonoid biosynthesis were marked (PDF 930.5 kb)
10725_2020_660_MOESM9_ESM.pdf
Supplementary Fig. 5 The phylogenetic tree of bHLH factors between C. morifolium and Arabidopsis thaliana. The cluster of factors annotated into AtEGL3, AtGL3 and TT8 regulating the anthocyanin and proanthocyanidin biosynthesis was marked (PDF 837.4 kb)
Rights and permissions
About this article
Cite this article
Wang, T., Yang, F., Guo, Q. et al. Long-read sequencing of Chrysanthemum morifolium transcriptome reveals flavonoid biosynthesis and regulation. Plant Growth Regul 92, 559–569 (2020). https://doi.org/10.1007/s10725-020-00660-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00660-x