Abstract
Bacterial sigma factor E (SigE) is a positive regulator of sugar catabolism via its interaction with the H subunit of Mg-chelatase (CHLH), which serves as both putative abscisic acid (ABA) receptor (ABAR) in Arabidopsis thaliana and positive regulator of strawberry fruit ripening. However, whether SigE influences strawberry fruit ripening has not been determined. We used RNA sequencing and a qPCR to confirm that the expression of the strawberry SigE gene (FaSigE) is rapidly upregulated in fruits turning red, suggesting FaSigE might mediate strawberry fruit ripening. Silencing FaSigE by intron-spliced hairpin RNA-mediated RNA interference significantly inhibited fruit ripening. This observation was confirmed by analyses of fruit firmness, soluble sugar, ABA, and anthocyanin contents, as well as transcript levels of genes related to fruit ripening and ABA signaling. Interestingly, a firefly luciferase complementation assay revealed that FaSigE can interact with FaABAR, while an in vitro fruit disc incubation test indicated ABA induces FaSigE expression. Moreover, a surface plasmon resonance assay proved that FaABAR produced in yeast cells can bind to ABA, with a binding dissociation constant of 50 µM. In conclusion, FaSigE can interact with FaABAR and positively regulates strawberry fruit ripening via ABA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past few decades, the hormone-regulated molecular mechanisms underlying the ripening of fleshy fruits have been studied extensively, with investigations confirming that ethylene has a key effect on climacteric fruits and abscisic acid (ABA) is important for non-climacteric fruits (Kumar et al. 2014; Shen and Joss 2014). Additionally, FaCHLH/ABAR (H subunit of Mg-chelatase/putative ABA receptor) positively regulates the ripening of strawberry fruits in response to ABA (Jia et al. 2011), while the interaction between CHLH and SigE (sigma factor E) is critical for the catabolism of sugar in Synechocystis species (Osanai et al. 2009). However, to the best of our knowledge, whether SigE is involved in the ripening of strawberry fruits has not been determined.
RNA polymerases play a central role in gene expression and regulation, and are essential for living organisms to adapt to various environmental and physiological conditions. Bacterial RNA polymerase contains four core subunits (RpoA, RpoB, RpoC1, and RpoC2) and nine sigma factors (SigA of group 1, SigB to SigE of group 2, and SigF to SigI of group 3) that bind to the RNA polymerase to initiate transcription in response to an environmental stimulus (Kaneko et al. 1996; Haugen et al. 2008; Imamura et al. 2003; Lemeille et al. 2005). Previous studies revealed that SigE, which is an extra cytoplasmic function sigma factor, is crucial for bacterial stress and inflammatory responses (Missiakas and Raina 1998; Helmann 2002; Staron et al. 2009; Manganelli and Provvedi 2010; Pátek and Nešvera 2011; Buasri and Panbangred 2012; Antal et al. 2016; Gupta et al. 2016). A poorly-conserved TTHB212 protein, which exhibits anti-SigE properties, inhibits transcription by preventing SigE from binding to RNA polymerase (Sakamoto et al. 2008). Interestingly, SigE regulates gene expression specifically during the post-exponential growth phase of Synechococcus species (Gruber and Bryant 1998). Thus, as a cell surface stress-related transcription factor, SigE is important for bacterial growth and stress resistance, with diverse effects on cell sporulation (Shuler et al. 1995; Wong et al. 1995; Al-Hinai et al. 2014; Casonato et al. 2014; Kirk et al. 2014), granuloma formation (Giacomini et al. 2006), phagocytosis by macrophages (Ando et al. 2003), cell wall integrity (Paget et al. 1999), fatty acid metabolism (Manganelli et al. 2001), carotenoid biosynthesis (Hakkila et al. 2013), nitrogen starvation and sugar catabolism (Osanai et al. 2005a), and responses to various stresses, including acid (Bansal et al. 2017), oxidative (Bagchi and Ghosh 2006; Pacheco et al. 2012), heat (Pettersson et al. 2015), and salt (Koskinen et al. 2016) stresses.
The PII signaling proteins help coordinate carbon and nitrogen metabolism in bacteria, archaea, and plant chloroplasts. In Synechocystis spp. PCC 6803, PamA is a PII-binding transmembrane protein that positively regulates the expression of genes involved in sugar catabolism via SigE (Osanai et al. 2005b, 2006, 2009, 2011; Summerfield et al. 2007; Azuma et al. 2011). Interestingly, the chloroplast H subunit of Mg-chelatase can interact with SigE to inhibit the transcriptional activity of SigE (Osanai et al. 2009). In fact, the complex regulation of SigE activity occurs at the transcriptional, translational, and post-translational levels. Specifically, the transcriptional regulation is mediated by several two-component systems including MprAB (He et al. 2006; Donà et al. 2008), CseB/CseC (Paget et al. 1999; Hong et al. 2002), and PhoP/PhoR (Prágai et al. 2004). Meanwhile, the post-translational regulation is mediated by RseA (Hutchings et al. 2006; Manganelli and Provvedi 2010) and CseE (Park et al. 2008).
Although the roles of SigE in bacteria have been relatively well studied, only a few functions of SigE in higher plants have been reported (Fujiwara et al. 2000). The downregulated expression of FaCHLH/ABAR increases the SigE transcript level in unripened strawberry fruits (Jia et al. 2011), while an interaction between CHLH and SigE inhibits the transcriptional activity of SigE related to sugar catabolism in Synechocystis species (Osanai et al. 2009). These findings compelled us to investigate the role of SigE during strawberry fruit ripening. We cloned a strawberry SigE homolog, FaSigE, and determined its effects on fruit ripening using intron-spliced hairpin RNA (ihpRNA; Hoffmann et al. 2006). Additionally, we clarified the relationships among FaSigE, FaABAR, and ABA by analyzing the interaction between FaABAR and FaSigE in a firefly luciferase complementation (LUC) assay, determining the effects of ABA on FaSigE transcript abundance during an in vitro fruit disc incubation test, and examining the binding of ABA by FaABAR based on surface plasmon resonance (SPR). Our results revealed that FaSigE interacts with FaABAR, and positively regulates strawberry fruit ripening via ABA.
Results
Cloning, bioinformatics, expression and function analysis of FaSigE in strawberry fruits
A BLAST search conducted using the Arabidopsis thaliana SigE (GenBank: AB021120.1) sequence from the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) as a query identified a highly homologous Fragaria vesca subsp. vesca gene (GenBank: XM_004307961) with a 1,981-bp full-length cDNA comprising a 1563-bp coding sequence (172–1,734 bp). Gene-specific primers (forward: 5′-ATGGGAGTTGTGACAGTTTC-3′; reverse: 5′-CAACTATGTATCTGCGAA G-3′) were designed for a subsequent amplification of the strawberry SigE coding sequence in a reverse transcription polymerase chain reaction (RT-PCR) using a cDNA library prepared from strawberry fruits. We then isolated the 1,563-bp amplified product (i.e., FaSigE; Fig. 1a), which included an open reading frame encoding a protein with 520 amino acids. The deduced protein contained a conserved domain with the sigma-70 factor, RpoD, which was detected during a homology analysis involving a BLAST search of the NCBI database (Fig. 1b). Thus, the putative strawberry FaSigE gene was cloned successfully.
Cloning, bioinformatics, expression and function analysis of FaSigE in strawberry fruits. a Agarose gel electrophoresis analysis of a 1563-bp FaSigE fragment from strawberry fruits generated by a reverse transcription polymerase chain reaction (PCR). M marker, 2000–100 bp. b Putative conserved domains (RNA polymerase sigma factor, RpoD-like family, cyanobacteria) were determined based on a 521-amino acid FaSigE sequence in the NCBI database (http://blast.ncbi.nlm.nih.gov/ Blast.cgi). The identified protein family was most closely related to the essential sigma-70 factor RpoD in the C-terminal domain model, including the sigma70_r2, sigma70_r3, and sigma70_r4 superfamilies. c Two SigE gene-like contigs were detected in the RNA sequencing transcriptome data for the fruits collected during the large green (LG), white (Wt), initial red (IR), and partial red (PR) stages. d Real-time PCR analysis of FaSigE expression in fruits collected during the LG, Wt, IR, and PR. e Control fruits infected with empty vector turned red. f The fruits in which FaSigE was silenced by RNA interference exhibited a chimeric phenotype. g Real-time PCR analysis of the FaSigE expression levels in transgenic and control fruits. Actin expression levels were used as the internal control. Error bars represent standard errors (n = 3). Significant differences determined by variance analysis followed by Duncan’s multiple range tests are indicated by different lowercase letters (P < 0.05). (Color figure online)
To analyze the FaSigE transcript profile during the onset of strawberry fruit ripening, four mixed cDNA libraries derived from ten uniformly-sized fruits at various stages, including large green (LG), white (Wt), initial red (IR), and partial red (PR) stages, underwent RNA-sequencing (RNA-seq) using the Illumina HiSeq 2000 instrument. Compared with the LG stage, which was associated with 709,101 unigenes, the Wt (537,267 unigenes), IR (534,780 unigenes), and PR (539,813 unigenes) stages had fewer and similar unigenes. The data indicated that the Wt stage represents the transition from green to red fruits, and is a distinct stage during the onset of fruit ripening. We detected two SigE-like contigs based on RNA-seq data. The expression levels of contig comp80129_c0_seq1 and its complementary strand, comp80826_c0_seq2, were high in large green fruits, but then decreased and then rapidly increased as fruits turned red (Fig. 1c). Thus, comp80129_c0_seq1 was considered to be FaSigE. This result was confirmed by real-time PCR (Fig. 1d). These results suggested that FaSigE might influence strawberry fruit ripening.
To clarify the role of FaSigE in ripening strawberry fruits, FaSigE was silenced in fruits using ihpRNA (Intron splicing hpRNA). First, degreening fruits were inoculated with Agrobacterium tumefaciens strain GV3101 cells transformed with the pH7FWG vector carrying the FaSigE-ihpRNA construct. Fruits treated with bacterial cells containing the empty vectors were used as control. Two-weeks later, the control fruits were completely red (Fig. 1e), unlike the inoculated region of RNAi-treated fruits (Fig. 1f). A qPCR analysis indicated that FaSigE expression levels were significantly lower in RNAi-treated fruits (mixed samples) than in control fruits (Fig. 1g). These results implied that FaSigE contributes to the development of red strawberry fruits at the molecular level.
Downregulation of FaSigE expression affects a set of ripening-related parameters in RNAi fruits
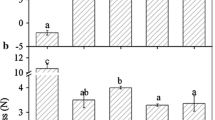
To further characterize FaSigE functions related to fruit ripening, we analyzed ripening-related physiological parameters, including firmness, soluble sugar concentration, anthocyanin content, and ABA level. Fruits in which FaSigE was silenced were firmer than the control fruits (Fig. 2a). Additionally, the anthocyanin (Fig. 2b), soluble sugar (Fig. 2c), and ABA (Fig. 2d) contents were lower in the RNAi-treated fruits. Furthermore, an investigation of the transcript abundance of genes related to ripening and ABA signaling, including FaCHS, FaUFGT, FaPG1, FaPL1, FaSUT1, FaAI, FaPYR1, and FaABI4 (Jia et al. 2011, 2013a), revealed that the expression levels of all genes were downregulated (Fig. 2e). Therefore, the differences between the control and RNAi-treated fruits regarding the expression of genes related to fruit firmness (FaPG1and FaPL1), sugar content (FaSUT1and FaAI), anthocyanin content (FaCHS and FaUFGT), and ABA signaling (FaPYR1 and FaABI4) implied that FaSigE is a positive regulator of strawberry fruit ripening.
Changes to several physiological parameters and fruit ripening- and ABA signaling-related gene expression in transgenic and control fruits. a Firmness. b Anthocyanin contents. c Soluble sugar (i.e., sucrose, glucose, and fructose) contents. d Abscisic acid (ABA) contents. e Relative expression levels of genes related to fruit ripening and ABA signaling. Error bars represent standard errors (n = 3). Significant differences determined by variance analysis followed by Duncan’s multiple range tests are indicated by different lowercase letters (P < 0.05)
FaSigE interacts with FaABAR that can bind ABA
Previous studies confirmed that FaCHLH/ABAR positively regulates strawberry fruit ripening (Jia et al. 2011) and Synechocystis SigE can interact with CHLH (Osanai et al. 2009), suggesting that the regulation of strawberry fruit ripening by FaSigE might be influenced by ABA. To verify this possibility, we first analyzed the interaction between FaSigE and FaABAR in a firefly LUC assay. Because the full-length ABAR/CHLH sequence (i.e., 1,381 amino acids) was too long to be an appropriate bait, we cloned a C-terminal ABAR fragment encoding 690 amino acids (ABARc690) that are sufficient for interacting with ABA and transmitting the ABA signal (Wu et al. 2009). The C-terminal ABAR fragment and SigE were inserted into the pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors, which were used to transform Nicotiana benthamiana cells. An imaging test indicated that ABARc690 could interact with SigE, unlike the control pairings with ABARc690-nLUC and cLUC or nLUC and SigE-cLUC (Fig. 3a). Second, we analyzed the effects of ABA on FaSigE expression levels using an in vitro fruit disc incubation test. The qPCR results indicated that the FaSigE transcript abundance was significantly higher in the fruit discs exposed to 100 µM ABA for 2 h than in the mannitol-treated control discs (Fig. 3b). These results implied that ABA induces FaSigE expression. Third, another qPCR assay confirmed that the FaABAR transcript level was considerably lower in FaSigE-silenced fruits (mixed samples) than in control fruits (Fig. 3c), suggesting that FaSigE might positively regulate FaABAR expression.
FaABAR interacts with FaSigE and can bind ABA. a Interaction between FaABAR and FaSigE based on a firefly luciferase complementation (LUC) assay. The ABARc690-nLuc and cLuc-SigE constructs were used to assess the interaction between FaABAR and FaSigE in a firefly LUC assay. The fluorescence signals at −110 °C, which were recorded with a 1300B CCD camera (Roper), confirmed that ABARc690 can interact with SigE, while interactions were not detected between ABARc690-nLUC and cLUC or nLUC and SigE-cLUC. b Real-time PCR analysis of FaABAR expression in FaSigE-silenced and control fruits. c Effects of abscisic acid on FaSigE transcript abundance in an in vitro fruit disc incubation test. Actin transcript abundance was used as the internal control. d Agarose gel electrophoresis analysis of the 2073-bp FaABAR/CHLH fragment from strawberry fruits generated by a reverse transcription polymerase chain reaction. M marker, 2000–100 bp. e Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of the recombinant protein (i.e., FaABARc690) produced in yeast cells. M marker, 100–25 kDa; 0–36 h, duration of the induction of FaABARc690 production with 1% methanol. f Purification of the recombinant protein using Ni–NTA resin and identification of FaABARc690 in a western blot. M marker, 100–25 kDa. g and h Surface plasmon resonance analysis of the binding of FaABARc690 to abscisic acid (ABA). The sample protein was immobilized at the surface of a sensor chip by an amino-coupling process. The binding of (+)-ABA to the immobilized proteins was analyzed based on response data. g Data of a representative response record. h Corresponding saturation curve of ABA binding to FaABARc690. Error bars represent standard errors (n = 3). Significant differences determined by variance analysis followed by Duncan’s multiple range tests are indicated by different lowercase letters (P < 0.05)
Although the role of FaABAR in ripening strawberry fruits has been determined (Jia et al. 2011), it remains unclear whether FaABAR can bind to ABA. Clarifying this point may help to elucidate the molecular mechanism underlying FaSigE activity. Consequently, we used SPR to assess the interaction between ABA and FaABAR. Because the C-terminal half of A. thaliana ABAR interacts with ABA (Wu et al. 2009), a cDNA fragment [i.e., 2104–4143 bp (GenBank No. GQ201451.1)] encoding a 680-amino acid C-terminal region was isolated using a strawberry cDNA library (Fig. 3d). The 2,040-bp FaABAR fragment was cloned into pPICZB, which was then linearized and inserted into Pichia pastoris X-33 cells. Isolated transformants were treated with 1% methanol to induce the production of a histidine-tagged FaABAR fusion protein. Over the course of a 36-h induction period, production of the 75-kDa recombinant protein initially increased gradually, peaking at 24 h (Fig. 3e). The histidine-tagged FaABAR was purified using the Ni–NTA His Bind Resin and then analyzed in a western blot using an anti-histidine tag antibody (Fig. 3f). The final purified protein solution (0.5 mg/ml) was 93.7% pure. Next, an SPR assay was completed using a Biacore T200 system equipped with a certified CM5 sensor chip with carboxyl groups on its surface. We immobilized FaABAR (0.5 mg/ml) to the chip surface at pH 4.0. The analyte with 6–102 µM (+)-ABA in 20 mM phosphate buffer (pH 7.4) was applied at a flow rate of 50 µl/min. Three independent ABA binding measurements confirmed that ABA can bind to strawberry FaABAR, with an average dissociation constant of 50 ± 1.98 μM (Fig. 3g, h).
Discussion
During the past few decades, much has been made toward understanding the action mechanisms of bacterial SigE (Peters et al. 1991; Goto-Seki et al. 1999; Hong et al. 2002; Paget et al. 1999; Prágai et al. 2004; Osanai et al. 2006, 2009; Donà et al. 2008; Sakamoto et al. 2008; Manganelli and Provvedi 2010). Although FaCHLH/ABAR is a positive regulator of strawberry fruit ripening (Jia et al. 2011), whether SigE is involved has not been determined.
In the present study, we prove that FaSigE regulates strawberry fruit ripening via ABA. We revealed that the change in fruit color from green to red coincides with a rapid increase in FaSigE transcript abundance, with the highest levels in ripened fruits (Fig. 1). We also observed that the downregulated FaSigE expression in de-greening fruits inhibits ripening (Figs. 1, 2). Additionally, FaSigE interacts with FaABAR, which can bind ABA (Fig. 3), and FaSigE expression is induced by ABA (Fig. 3). Furthermore, ABA contents and FaABAR expression levels are downregulated in fruits in which FaSigE is silenced by RNAi (Figs. 2, 3).
It is notable that CHLH negatively regulates SigE transcription in Synechocystis sp. PCC 6803 (Osanai et al. 2009). Moreover, downregulated CHLH expression in unripened strawberry fruits promotes SigE transcription, but inhibits the accumulation of soluble sugars (Jia et al. 2011). Considering that Synechocystis species SigE positively regulates sugar catabolism (Azuma et al. 2011), it is possible that the inhibited accumulation of soluble sugars is because the increased transcription of SigE in unripened fruits results in excessive metabolism of sugars (Jia et al. 2011).
In the present study, the FaSigE transcript abundance increased rapidly during the strawberry fruit ripening process (Fig. 1), suggesting that FaSigE might be a positive regulator of ripening. A decrease in FaSigE transcripts may inhibit ripening and sugar accumulation in transgenic fruits (Figs. 1, 2), implying FaSigE positively influences sugar catabolism. Additionally, the inhibition of SigE expression downregulates FaABAR transcription and ABA levels, as a result, inhibits the fruit ripening (Figs. 2, 3). Our data suggest that SigE, FaABAR, sugars, and ABA are associated with a complex regulatory signaling network affecting strawberry fruit ripening. Given important roles of ABA and sugar in ripening (Han et al. 2015; Wang et al. 2015; Razzaq et al. 2016; Zhao et al. 2017), future studies should investigate SigE-related two-component systems and characterize the molecular mechanism that enables SigE to regulate strawberry fruit ripening via ABA signaling pathway.
Materials and methods
Plant materials
Octaploid strawberry (Fragaria ananassa cv. ‘Hongyan’) plants were cultivated on ridges of loam soil with 0.2 m × 0.3 m row spacing in a glasshouse (20–25 °C, 70–85% relative humidity, and a 14-h light/10-h dark cycle) during spring in 2014–2016. One acre (667 m2) of soil contained 5000 kg organic fertilizer and 30 kg synthetic fertilizer (i.e., nitrogen, phosphorus, and potassium). Plants were irrigated every 5–7 days.
We tagged 200 small green fruits on 50 strawberry plants after flowering. Forty uniformly-sized fruits were collected at 15 (LG), 23 (Wt), 25 (IR), 27 (PR), and 30 (FR) days after anthesis, quickly frozen in liquid nitrogen, and stored at −80 °C until analyzed.
RNA sequencing and data analysis
Total RNA was extracted from the receptacles of six frozen fruits per analyzed stage (i.e., LG, Wt, IR, and PR) using the RNeasy Plant Mini kit (Qiagen, Dusseldorf, Germany). The extracted RNA was treated with RNase-free DNase (Qiagen) to remove contaminating genomic DNA and then used as the template to generate cDNA libraries with the RNA Library Prep kit (New England BioLabs, Ipswich, MA, USA). The prepared libraries were sequenced using the Illumina HiSeq 2000 platform (Illumina, USA). The data analysis was done according to the description (Benjamini and Yekutieli 2001; Mortazavi et al. 2008; Wang et al. 2010, 2017; Langmead and Salzberg 2012; Ming et al. 2012).
RNA isolation and cDNA synthesis
Total RNA was extracted from 0.5 g strawberry receptacles from three frozen fruits of each stage using the Easyspin Rapid Plant RNA Extraction kit (Biomed, Beijing, China). The purity and integrity of the extracted RNA were analyzed by agarose gel electrophoresis and absorbance ratios (i.e., 260:230 nm and 260:280 nm), after which 3 µg RNA was reverse transcribed using the TransScript First-strand cDNA Synthesis SuperMix kit (Biomed).
Cloning of the FaSigE coding sequence
The cDNA obtained as described in the previous section was used as the template for amplifying FaSigE with gene-specific primers (forward, 5′-ATGGGAGTTGTGACAGTTTC-3′; reverse, 5′-TCAACTATGTATCTGCGAAG-3′) designed based on a sequence in a strawberry gene library database (https://strawberry.plantandfood.co.nz/index.html). The PCR program used to amplify the FaSigE sequence was as follows: 94 °C for 5 min; 34 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 90 s; 72 °C for 10 min. The PCR products were ligated into a pEASY-T1 simple vector for a subsequent transformation of Escherichia coli DH5α cells.
Construction of the FaSigE RNA interference vector
For the ihpRNA-mediated silencing of FaSigE, a 461-bp FaSigE cDNA fragment was amplified using the following primers: forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCCTCCTGTTCTTAGTTCCTCTG-3′ (attB1, underlined); reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGGATTTCTGCCCTGACC-3′ (attB2, underlined). The 461-bp amplified fragment was cloned into the pDONR™ 221 vector (Invitrogen) using the Gateway BP Clonase II enzyme mix, and into the pK7GWIWG2(II)-RR vector using the Gateway LR Clonase II enzyme mix. The FaSigE-RNAi recombinant plasmid was inserted into A. tumefaciens strain GV3101 cells, which were grown at 28 °C in Luria–Bertani liquid medium containing 10 mM MES, 20 µM acetosyringone. When the culture reached an optical density at 600 nm (OD600) of 0.8, the cells were harvested and resuspended in infection buffer [10 mM MgCl2, 10 mM MES (pH 5.6), and 200 µM acetosyringone]. The cell suspension was mixed for 2 h at room temperature before 200-µl aliquots were injected into whole fruits using a sterile 1 ml hypodermic syringe.
Determination of anthocyanin and soluble sugar contents
The anthocyanin content of three fruits of each stage was measured by reversed-phase high-performance liquid chromatography (HPLC) using a ZORBAX Eclipse XDB-C18 column (4.6 × 150 mm, 5 µm; Agilent) with the following linear gradient: Solution A (acetonitrile) 0 to 20% for 13 min, 20–40% for 20 min, and 0% for 25 min and Solution B (10% formic acid) at a flow rate of 1 ml/min. The detection wavelength was 520 nm, the column temperature was 25 °C, and the injection volume was 20 µl. Pelargonidin-3-O-glucoside was used as the standard. The HPLC-analysis of soluble sugar content was done according to the description (Jia et al. 2011). The analysis was repeated three times.
Quantitative real-time polymerase chain reaction
The qRT-PCR analyses were completed in a CFX Sequence Detector (Bio-Rad, Hercules, CA, USA) using 20-μl samples consisting of 10 μl SYBR Premix Ex Taq (Takara), 0.4 μl forward and reverse primers, and 2 μl cDNA template. The PCR program was as follows: 95 °C for 2 min; 40 cycles of 94 °C for 20 s and 54 °C for 30 s. Actin was used as a reference gene. Relative gene expression levels were analyzed using the 2−ΔΔCt method (Livak and Schmittgen 2001). The following qRT-PCR primers were used in Table 1.
Firefly luciferase complementation assay
To analyze the interaction between ABAR and SigE, we generated the ABARc690-nLuc and cLuc-SigE constructs. The pCAMBIA1300-nLUC and pCAMBIA1300-cLUC plasmids were kindly provided by Dr. Gong Zhi-Zhong of China Agricultural University. All constructs were inserted into A. tumefaciens strain GV3101 cells, which were subsequently used to transform Nicotiana benthamiana plants as follows. A single clone of each strain was cultured in 5 ml YEB supplemented with 50 mg/l kanamycin and 50 mg/l rifampicin at 28 °C for 24 h. The bacteria were then collected and re-suspended with 3 ml injection buffer 10 mM MES (pH 5.7), 10 mM MgCl2, and 250 mM acetosyringone for an OD600 of 0.5. The bacterial solutions were kept at room temperature for 2 h before being injected into N. benthamiana leaves. Three days later, 1 mM luciferin was sprayed onto the lower epidermis, after which the leaves were maintained in darkness for 5 min. The fluorescence signals were observed and recorded at −110 °C using a 1300B CCD camera (Roper). The assay was completed with three replicates.
Determination of firmness and abscisic acid content
Three uniform strawberry fruits of each stage were used to assess flesh firmness. Two sides of each fruit were analyzed using a GY-4 fruit penetrometer (Digital Force Gauge, Shanghai, China) and flesh firmness was recorded as N/cm2. The experiment was completed with three replicates.
To extract ABA, 0.5 g receptacles were ground using a mortar and pestle and homogenized in extraction solution [80% (v/v) methanol]. Extracts were centrifuged at 10,000×g for 20 min and the supernatants were added to Sep-Pak C18 cartridges (Waters, Milford, MA, USA) to remove polar compounds. Samples were stored at −20 °C prior to being used in an enzyme-linked immunosorbent assay. The experiment was completed with three replicates.
In vitro fruit disc incubation test
White strawberry fruit disc tissues were treated with ABA in an in vitro incubation test as described by Jia et al. (2013b). The discs were divided into two sections, with one section incubated in equilibration buffer with 200 mM mannitol (i.e., control) and the other section incubated in equilibration buffer with 100 µM ABA. The two sections were kept in a 250-ml flask at 25 °C for 2 h with shaking. The sections were washed with double-distilled water and then frozen in liquid nitrogen and kept at −80 °C. The experiment was completed with three replicates.
Cloning of the FaABAR abscisic acid-binding region
The cDNA generated as described above was used as the template for amplifying the C-terminal half of FaABAR with the following primers: forward, 5′-GAATTCAAAAATGGCTGCCAACAATCCATCTG-3′ (EcoRI, underlined); reverse, 5′-GCGGCCGCCCGATCGATTCCCTCAATTTTG-3′ (Not I, underlined). The PCR program was as follows: 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 130 s; 72 °C for 10 min. The amplification products were ligated into a pEASY-T1 simple vector and inserted into E. coli DH5α cells (TransGen Biotech). The plasmid DNA from positive colonies were amplified and sequenced by Invitrogen (Shanghai, China).
Purification of recombinant FaABAR produced in yeast cells
Recombinant FaABAR was expressed and purified using the EasySelect™ Pichia Expression Kit and the ProBond™ Purification System (Invitrogen, USA), respectively. A partial FaABAR sequence of N terminal was removed from a pEASY-T1 simple vector carrying FaABAR 2073 by digesting with EcoRI and NotI. The fragment was then cloned into the expression vector, pPICZB, digested with the same enzymes. The recombinant pPICZB plasmid was inserted into E. coli cells and transformants were selected in Petri dishes containing agar-solidified low-salt Luria–Bertani medium supplemented with 25 μg/ml Zeocin™. Twenty transformants were selected for sequencing to confirm the target fragment was inserted correctly (i.e., in frame with the C-terminal tag). The purified and linearized recombinant plasmids were inserted into P. pastoris cells and Zeocin™-resistant yeast transformants were selected in Petri dishes consisting of agar-solidified YPDS medium supplemented with an appropriate Zeocin™ concentration. The FaABAR fusion protein was expressed and purified using the Ni–NTA His Bind Resin and analyzed in a western blot involving an anti-histidine tag antibody according to manufacturer-recommended protocols (Novagen, USA). The eluted fusion protein was stored at −80 °C until analyzed further.
Surface plasmon resonance assay
The SPR assay was completed using a Biacore T200 system (GE Healthcare) equipped with a certified CM5 sensor chip with carboxyl groups on its surface. Sample proteins (90% pure based on size exclusion chromatography) were covalently immobilized to saturate the sensor chip surface using the Amine Coupling kit (Biacore). The surface of flow cell 2 was activated for 7 min with a 1:1 mixture of 0.1 M N-hydroxysuccinimide and 0.1 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride at a flow rate of 10 µl/min. The FaABAR (0.5 mg/ml in 10 mM sodium acetate, pH 4.0) was immobilized on the sensor chip surface at a density that saturated the surface. Meanwhile, flow cell 1 was left blank to serve as a control. The chip surface was then blocked with a 7 min injection of 1 M ethanolamine, pH 8.0. To collect kinetic and affinity binding data at 25 °C, the analyte consisting of (+)-ABA in 20 mM phosphate buffer (150 mM NaCl and 30 mM KCl, pH 7.4) was injected over flow cells 1 and 2 at 5–120 µM and a flow rate of 50 µl/min. The complex was allowed to associate and dissociate for 60 s, and data were collected and globally fitted to a steady-state model in the Biacore S200 Evaluation Software v1.0. The experiment was completed with three replicates.
Abbreviations
- SigE:
-
Sigma factor E
- ABA:
-
Abscisic acid
- CHLH:
-
The H subunit of Mg-chelatase
- ABAR:
-
Putative ABA receptor
- RNA-seq:
-
RNA sequence
- LUC:
-
Firefly luciferase complementation assay
- SPR:
-
Surface plasmon resonance
- ihpRNA:
-
Intron splicing hpRNA
References
Al Hinai MA, Jones SW, Papoutsakis ET (2014) σK of clostridium acetobutylicum is the first known sporulation-specific sigma factor with two developmentally separated roles, one early and one late in sporulation. J Bacteriol 196:287–299
Ando M, Yoshimatsu T, Ko C, Converse PJ, Bishai WR (2003) Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect Immun 71:7170–7172
Antal T, Kurkela J, Parikainen M, Kårlund A, Hakkila K, Tyystjärvi E, Tyystjärvi T (2016) Roles of group 2 sigma factors in acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to nitrogen deficiency. Plant Cell Physiol 57:1309–1318
Azuma M, Osanai T, Hirai MY, Tanaka K (2011) A response regulator Rre37 and an RNA polymerase sigma factor SigE represent two parallel pathways to activate sugar catabolism in a cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 52:404–412
Bagchi A, Ghosh TC (2006) Structural study of two proteins SigE and ORF1 to predict their roles in the biochemical oxidation of sulfur anions via the global sulfur oxidation operon (sox). Comput Biol Chem 30:227–232
Bansal R, Anil Kumar V, Sevalkar RR, Singh PR, Sarkar D (2017) Mycobacterium tuberculosis virulence-regulator PhoP interacts with alternative sigma factor SigE during acid-stress response. Mol Microbiol. doi:10.1111/mmi.13635
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Buasri W, Panbangred W (2012) Large crystal toxin formation in chromosomally engineered Bacillus thuringiensis subsp. aizawai due to σE accumulation. Appl Environ Microbiol 78:1682–1691
Casonato S, Provvedi R, Dainese E, Palù G, Manganelli R (2014) Mycobacterium tuberculosis requires the ECF sigma factor SigE to arrest phagosome maturation. PLoS ONE 9:e108893
Donà V, Rodrigue S, Dainese E, Palù G, Gaudreau L, Manganelli R, Provvedi R (2008) Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor sigmaE in Mycobacterium tuberculosis. J Bacteriol 190:5963–5971
Fujiwara M, Nagashima A, Kanamaru K, Tanaka K, Takahashi H (2000) Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Aarabidopsis thaliana. FEBS Lett 481:47–52
Giacomini E, Sotolongo A, Iona E, Severa M, Remoli ME, Gafa V, Lande R, Fattorini L, Smith I, Manganelli R, Coccia EM (2006) Infection of human dendritic cells with a Mycobacterium tuberculosis sigE mutant stimulates production of high levels of interleukin-10 but low levels of CXCL10: impact on the T-cell response. Infect Immun 74:3296–3304
Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H (1999) Specificity crosstalk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC7942: in vitro specificity and a phylogenetic analysis. Mol Microbiol 34:473–448
Gruber TM, Bryant DA (1998) Characterization of the alternative sigma-factors SigD and SigE in Synechococcus sp. strain PCC 7002. SigE is implicated in transcription of post-exponential-phase-specific genes. Arch Microbiol 169:211–219
Gupta AM, Pal P, Mandal S (2016) Structural analysis of sigma E interactions with core RNA polymerase and its cognate P-hsp20 promoter of Mycobacterium tuberculosis. J Biomol Struct Dyn 34:792–799
Hakkila K, Antal T, Gunnelius L, Kurkela J, Matthijs HC, Tyystjärvi E, Tyystjärvi T (2013) Group 2 sigma factor mutant ∆sigCDE of the cyanobacterium Synechocystis sp. PCC 6803 reveals functionality of both carotenoids and flavodiiron proteins in photoprotection of photosystem II. Plant Cell Physiol 54:1780–1790
Han Y, Dang R, Li J, Jiang J, Zhang N, Jia M, Wei L, Li Z, Li B, Jia W (2015) SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol 167:915–930
Haugen SP, Ross W, Gourse RL (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6:507–519
He H, Hovey R, Kane J, Singh V, Zahrt TC (2006) MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol 188:2134–2143
Helmann JD (2002) The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110
Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48:818–826
Hong HJ, Paget MS, Buttner MJ (2002) A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol Microbiol 44:1199–1211
Hutchings MI, Hong HJ, Leibovitz E, Sutcliffe IC, Buttner MJ (2006) The sigma(E) cell envelope stress response of Streptomyces coelicolor is influenced by a novel lipoprotein, CseA. J Bacteriol 188:7222–7229
Imamura S, Yoshihara S, Nakano S, Shiozaki N, Yamada A, Tanaka K, Takahashi H, Asayama M, Shirai M (2003) Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J Mol Biol 325:857–872
Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157:188–199
Jia HF, Lu D, Sun JH, Li CL, Xing Y, Qin L, Shen YY (2013a) Type 2 C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot 64:1677–1687
Jia HF, Li CL, Chai YM, Xing Y, Shen YY (2013b) Sucrose promotes strawberry fruit ripening by stimulation of abscisic acid biosynthesis. Pak J Bot 45:169–175
Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3:109–136
Kirk DG, Zhang Z, Korkeala H, Lindström M (2014) Alternative sigma factors SigF, SigE, and SigG are essential for sporulation in Clostridium botulinum ATCC 3502. Appl Environ Microbiol 80:5141–5150
Koskinen S, Hakkila K, Gunnelius L, Kurkela J, Wada H, Tyystjärvi T (2016) In vivo recruitment analysis and a mutant strain without any group 2 σ factor reveal roles of different σ factors in cyanobacteria. Mol Microbiol 99:43–54
Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65:4561–4575
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lemeille S, Latifi A, Geiselmann J (2005) Inferring the connectivity of a regulatory network from mRNA quantification in Synechocystis PCC6803. Nucleic Acids Res 33:3381–3389
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods 25:402–408
Manganelli R, Provvedi R (2010) An integrated regulatory network including two positive feedback loops to modulate the activity of sigma (E) in mycobacteria. Mol Microbiol 75:538–542
Manganelli R, Voskuil MI, Schoolnik GK, Smith I (2001) The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437
Manganelli R, Fattorini L, Tan D, Iona E, Orefici G, Altavilla G, Cusatelli P, Smith I (2004) The extra cytoplasmic function sigma factor sigma(E) is essential for Mycobacterium tuberculosis virulence in mice. Infect Immun 72:3038–3041
Ming YL, Gui RQ, Jing J, Hui QY, Li HX, Jin ZX, Ren YZ (2012) Transcriptome sequencing and de novo analysis for bamboo using the illumina platform. PLoS ONE 7:1–11
Missiakas D, Raina S (1998) The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol 28:1059–1066
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Method 5:621–628
Osanai T, Kanesaki Y, Nakano T, Takahashi H, Asayama M, Shirai M, Kanehisa M, Suzuki I, Murata N, Tanaka K (2005a) Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor SigE. J Biol Chem 280:30653–30659
Osanai T, Sato S, Tabata S, Tanaka K (2005b) Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J Biol Chem 280:34684–34690
Osanai T, Imamura S, Asayama M, Shirai M, Suzuki I, Murata N, Tanaka K (2006) Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res 13:185–195
Osanai T, Imashimizu M, Seki A, Sato S, Tabata S, Imamura S, Asayama M, Ikeuchi M, Tanaka K (2009) ChlH, the H subunit of the Mg-chelatase, is an anti-sigma factor for SigE in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 106:6860–6865
Osanai T, Oikawa A, Azuma M, Tanaka K, Saito K, Hirai MY, Ikeuchi M (2011) Genetic engineering of group 2 σ factor SigE widely activates expressions of sugar catabolic genes in Synechocystis sp. PCC 6803. J Biol Chem 286:30962–30971
Pacheco LG, Castro TL, Carvalho RD, Moraes PM, Dorella FA, Carvalho NB, Slade SE, Scrivens JH, Feelisch M, Meyer R, Miyoshi A, Oliveira SC, Dowson CG, Azevedo V (2012) A role for Sigma Factor σ(E) in Corynebacterium pseudotuberculosis Resistance to Nitric Oxide/Peroxide Stress. Front Microbiol. 3:126
Paget MS, Leibovitz E, Buttner MJ (1999) A putative two-component signal transduction system regulates sigmaE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2). Mol Microbiol 33:97–107
Park SD, Youn JW, Kim YJ, Lee SM, Kim Y, Lee HS (2008) Corynebacterium glutamicum sigmaE is involved in responses to cell surface stresses and its activity is controlled by the anti-sigma factor CseE. Microbiology 154:915–923
Pátek M, Nešvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154:101–113
Peters HK 3rd, Haldenwang WG (1991) Synthesis and fractionation properties of SpoIIGA, a protein essential for pro-sigma E processing in Bacillus subtilis. J Bacteriol 173:7821–7827
Pettersson BM, Das S, Behra PR, Jordan HR, Ramesh M, Mallick A, Root KM, Cheramie MN, de la Cruz Melara I, Small P, Dasgupta S, Ennis DG, Kirsebom LA (2015) Comparative Sigma Factor-mRNA Levels in Mycobacterium marinum under stress conditions and during host infection. PLoS ONE. 10:e0139823
Prágai Z, Allenby NE, O’Connor N, Dubrac S, Rapoport G, Msadek T, Harwood CR (2004) Transcriptional regulation of the phoPR operon in Bacillus subtilis. J Bacteriol 186:1182–1190
Razzaq K, Singh Z, Khan AS, Khan SAKU, Ullah S (2016) Role of 1-MCP in regulating ‘Kensington Pride’ mango fruit softening and ripening. Plant Growth Regul 78:401–411
Sakamoto K, Agari Y, Yokoyama S, Kuramitsu S, Shinkai A (2008) Functional identification of an anti-sigmaE factor from Thermus thermophilus HB8. Gene 423:153–159
Shen YY, Rose JKC (2014) ABA metabolism and signaling in fleshy fruits. In: Zhang DP (ed) Abscisic acid: metabolism, transport and signaling, Springer, Dordrecht, pp 271–286
Shuler MF, Tatti KM, Wade KH, Moran CP Jr (1995) A single amino acid substitution in sigma E affects its ability to bind core RNA polymerase. J Bacteriol 177:3687–3694
Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74:557–581
Summerfield TC, Sherman LA (2007) Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 189:7829–7840
Wang Z, Fang B, Chen J, Zhang X, Luo Z, Huang L, Chen X, Li Y (2010) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of SSR markers in sweet potato (Ipomoea batatas). BMC Genom 11:726
Wang YP, Chai P, Sin L, Li Q, Dai SJ, Sun YF, Kai WB, Zhang YS, Liang B, Leng P (2015) Transcriptional regulation of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and in response to abscisic acid and auxin at onset of fruit ripening. Plant Growth Regul 75:455–464
Wang QH, Zhao C, Zhang M, Li YZ, Shen YY, Guo JX (2017) Transcriptome analysis around the onset of strawberry fruit ripening uncovers an important role of oxidative phosphorylation in ripening. Sci Rep 7:41477
Wong J, Sass C, Bennett GN (1995) Sequence and arrangement of genes encoding sigma factors in Clostridium acetobutylicum ATCC 824. Gene 153:89–92
Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C, Zhao CX, Wang XF, Shang Y, Jiang T, Zhang XF, Yan L, Zhao R, Cui ZN, Liu R, Sun HL, Yang XL, Su Z, Zhang DP (2009) The Mg-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150:1940–1954
Zhao C, Hua LN, Liu XF, Li YZ, Shen YY, Guo JX (2017) Sucrose synthase FaSS1 plays an important role in the regulation of strawberry fruit ripening. Plant Growth Regul 81:175–181
Acknowledgements
This work was supported by the China National Science Foundation (Project 31672125), the Beijing Natural Science Foundation (6171001), Beijing Training Project for the Leading Talents in S & T (Grant No. LJ201612), and Beijing Municipal Education Commission (Grant No. CEFF-PXM2016-014207-000038).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhang, S., Hou, B., Chai, L. et al. Sigma factor FaSigE positively regulates strawberry fruit ripening by ABA. Plant Growth Regul 83, 417–427 (2017). https://doi.org/10.1007/s10725-017-0308-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-017-0308-x