Abstract
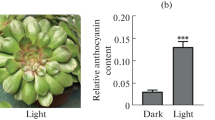
Anthocyanin biosynthesis in herbaceous plants has been studied widely, but the mechanism of anthocyanin accumulation in ‘Zijuan’ tea remains unclear. In this study, the leaf transcriptome of ‘Zijuan’ tea was analyzed at the purple and green stages by using the Illumina HiSeq 2500 platform. A total of 2250 unigenes were differentially expressed at the purple and green stages. Anthocyanin biosynthesis and carbohydrate metabolism were enriched through the KEGG metabolic pathway analysis of the differentially expressed unigenes. The upregulated sucrose synthase gene in the purple leaves may provide hexoses and UDP-glucose for anthocyanin synthesis. The genes encoding the corresponding enzymes in the glycolysis pathway were upregulated, whereas the expression of those genes encoding the enzymes in the TCA cycle showed no obvious change. Increased acetyl-CoA may contribute to anthocyanin accumulation through the upregulated cytosolic acetyl-coenzyme A carboxylase (ACCase) gene in the purple leaves of ‘Zijuan’ tea, indicating that glycolysis may provide more intermediates as substrates for promoting anthocyanin accumulation. The physiological and qRT-PCR results were consistent with the RNA-Seq data. Our findings expand our knowledge on the metabolic regulation of anthocyanin synthesis in tea plants.





Similar content being viewed by others
References
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119
Baroja-Fernández E, Muñoz FJ, Li J, Bahaji A, Almagro G, Montero M, Etxeberria E, Hidalgo M, Sesma MT, Pozueta-Romero J (2012) Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc Natl Acad Sci USA 109:321–326
Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122(1):107–116
Botstein D, Cherry J, Ashburner M, Ball C, Blake J, Butler H, Davis A, Dolinski K, Dwight S, Eppig J (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Cheng GW, Breen PJ (1991) Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J Am Soc Hortic Sci 116:865–869
Clark ST, Verwoerd WS (2011) A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinform 12:343
Conn S, Curtin C, Bézier A, Franco C, Zhang W (2008) Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Exp Bot 59:3621–3634
Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM (2003) Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131(3):259–268
De Jong W, Eannetta N, De Jong D, Bodis M (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108:423–432
Dean JV, Mohammed LA, Fitzpatrick T (2005) The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221:287–296
Debeaujon I, Peeters AJ, Léon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13:853–871
Deng Y, Li J, Wu S, Zhu Y, Chen Y, He F (2006) Integrated nr database in protein annotation system and its localization. Comput Eng 32:71–76
Fan JP, Fan C, Dong WM, Gao B, Yuan W, Gong JS (2013) Free radical scavenging and anti-oxidative activities of an ethanol-soluble pigment extract prepared from fermented Zijuan Pu-erh tea. Food Chem Toxicol 59:527–533
Fang CX, He HB, Wang QS, Qiu L, Wang HB, Zhuang YE, Xiong J, Lin WX (2010) Genomic analysis of allelopathic response to low nitrogen and barnyardgrass competition in rice (Oryza sativa L.). Plant Growth Regul 61(3):277–286
Fatland BL, Ke J, Anderson MD, Mentzen WI, Cui LW, Allred CC, Johnston JL, Nikolau BJ, Wurtele ES (2002) Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol 130:740–756
Fukusaki E, Kawasaki K, Kajiyama S, Kajiyama S, An C, Suzuki K, Tanaka Y, Kobayashi A (2004) Flower color modulations of Torenia hybrida by downregulation of chalcone synthase genes with RNA interference. J Biotechnol 111(3):229–240
Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549
Gong J, Zhang Q, Peng C, Fan J, Dong W (2012) Curie-point pyrolysis-gas chromatography-mass spectroscopic analysis of theabrownins from fermented Zijuan tea. J Anal Appl Pyrol 97:171–180
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Guay C, Madiraju SM, Aumais A, Joly É, Prentki M (2007) A role for ATP-citrate lyase, malic enzyme, and pyruvate/citrate cycling in glucose-induced insulin secretion. J Biol Chem 282:35657–35665
Guo J, Wang MH (2010) Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul 62(1):1–8
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071
Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126:1139–1149
Jia ZS, Tang MC, Wang JM (1999) The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem 64(4):555–559
Jia H, Wang JA, Yang Y, Liu G, Bao Y, Cui H (2015) Changes in flavonol content and transcript levels of genes in the flavonoid pathway in tobacco under phosphorus deficiency. Plant Growth Regul 76(2):225–231
Jiang L, Shen X, Shoji T, Kanda T, Zhou J, Zhao L (2013) Characterization and activity of anthocyanins in Zijuan tea (Camellia sinensis var. kitamura). J Agric Food Chem 61:3306–3310
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280
Kim S, Binzel M, Park S, Yoo KS, Pike L (2004) Inactivation of DFR (dihydroflavonol 4-reductase) gene transcription results in blockage of anthocyanin production in yellow onions (Allium cepa). HortScience 39(4):889–889
Kim JS, Lee BH, Kim SH, Oh KH, Cho KY (2006) Responses to environmental and chemical signals for anthocyanin biosynthesis in non-chlorophyllous corn (Zea mays L.) leaf. J Plant Biol 49:16–25
Koes RE, Spelt CE, van den Elzen PJ, Mol JN (1989) Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81:245–257
Larronde F, Krisa S, Decendit A, Cheze C, Deffieux G, Mérillon J (1998) Regulation of polyphenol production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Rep 17:946–950
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29:1035–1043
Leonard E, Lim KH, Saw PN, Koffas MA (2007) Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microb 73:3877–3886
Li W, Wang B, Wang M, Chen M, Yin JM, Kaleri GM, Zhang RJ, Zuo TN, You X, Yang Q (2014) Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J Integr Plant Biol 56:364–372
Markham KR, Ofman DJ (1993) Lisianthus flavonoid pigments and factors influencing their expression in flower colour. Phytochemistry 34:679–685
Mondal TK, Bhattacharya A, Laxmikumaran M, Ahuja PS (2004) Recent advances of tea (Camellia sinensis) biotechnology. Plant Cell Tiss Org 76:195–254
Ragg H, Kuhn DN, Hahlbrock K (1981) Coordinated regulation of 4-coumarate:CoA ligase and phenylalanine ammonia-lyase mRNAs in cultured plant cells. J Biol Chem 256(19):10061–10065
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25
Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57:675–709
Sasaki Y, Nagano Y (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotech Bioch 68:1175–1184
Sato M, Kawabe T, Hosokawa M, Tatsuzawa F, Doi M (2011) Tissue culture-induced flower-color changes in Saintpaulia caused by excision of the transposon inserted in the flavonoid 3′, 5′ hydroxylase (F3′ 5′ H) promoter. Plant Cell Rep 30(5):929–939
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646
Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15:462–470
Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Philos Trans R Soc Lond B Biol Sci 368:20120432
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
Van Der Krol AR, Mur LA, de Lange P, Mol JNM, Stuitje AR (1990) Inhibition of flower pigmentation by antisense CHS genes: promoter and minimal sequence requirements for the antisense effect. Plant Mol Biol 14(4):457–466
Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Mérillon JM (2000) Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry 53:659–665
Voll LM, Hajirezaei MR, Czogalla-Peter C, Lein W, Stitt M, Sonnewald U, Börnke F (2009) Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol 184:607–618
Wang QP, Peng CX, Gao B, Gong JS (2012) Influence of large molecular polymeric pigments isolated from fermented Zijuan tea on the activity of key enzymes involved in lipid metabolism in rat. Exp Gerontol 47:672–679
Wang ZW, Jiang C, Wen Q, Wang N, Tao YY, Xu LA (2014) Deep sequencing of the Camellia chekiangoleosa transcriptome revealed candidate genes for anthocyanin biosynthesis. Gene 538:1–7
Wind J, Smeekens S, Hanson J (2010) Sucrose: metabolite and signaling molecule. Phytochemistry 71:1610–1614
Xie L, Wang ZH, Cheng XH, Gao JJ, Zhang ZP, Wang LJ (2013) 5-Aminolevulinic acid promotes anthocyanin accumulation in Fuji apples. Plant Growth Regul 69(3):295–303
Yamamura Y, Ogihara Y, Mizukami H (2001) Cinnamic acid 4-hydroxylase from Lithospermum erythrorhizon: cDNA cloning and gene expression. Plant Cell Rep 20(7):655–662
Yang S, Zhang XN, Lu GL, Wang CR, Wang R (2015) Regulation of gibberellin on gene expressions related with the lignin biosynthesis in ‘Wangkumbae’pear (Pyrus pyrifolia Nakai) fruit. Plant Growth Regul 76(2):127–134
Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580:1183–1191
Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:17
Zheng X, Tian S (2006) Effect of oxalic acid on control of postharvest browning of litchi fruit. Food Chem 96:519–523
Acknowledgments
This work was financially supported by the Specialized Research Fund for the Doctoral Program of Higher Education of China (Award No. 20123515110014) and Natural Science Foundation of Fujian Province, China (Award No. 2014J01079).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
W. Chen is the first corresponding author.
J. Li and X. Lv have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2016_183_MOESM1_ESM.tif
Supplementary Fig. S1 Length distribution of contig and unigene sequence in the ‘Zijuan’ tea transcriptome. Length distribution of contigs(A); length distribution of unignes(B) (TIF 403 KB)
10725_2016_183_MOESM4_ESM.xlsx
Supplementary Table S3 Significantly enriched KEGG pathways for the differentially expressed unigenes in the ‘Zijuan’ tea transcriptome (XLSX 11 KB)
10725_2016_183_MOESM5_ESM.xlsx
Supplementary Table S4 Differentially expressed unigenes participated in anthocyanin biosynthesis and transportation and carbohydrate metabolism and some unigenes involved in TCA cycle between purple and green leaves in ‘Zijuan’ tea (XLSX 30 KB)
Rights and permissions
About this article
Cite this article
Li, J., Lv, X., Wang, L. et al. Transcriptome analysis reveals the accumulation mechanism of anthocyanins in ‘Zijuan’ tea (Camellia sinensis var. asssamica (Masters) kitamura) leaves. Plant Growth Regul 81, 51–61 (2017). https://doi.org/10.1007/s10725-016-0183-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0183-x