Abstract
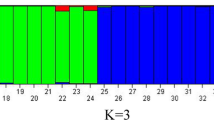
Molecular characterization and genetic diversity analysis were performed using SSR molecular markers in a commercially important ornamental chrysanthemum (Dendranthema grandiflora Tzvelev) involving standard cultivars and newly evolved genotypes. The total number of alleles was found to be 113 with an average of 4.34 per locus using 26 polymorphic SSR primers out of the 35 total SSRs initially screened. An average of 90.53 percent polymorphism was observed in the characterized genotypes with an average number of 1.03 monomorphic and 3.31 polymorphic bands. The mean polymorphic information content, effective multiplex ratio (EMR), Shannon index (I), expected (He), observed heterozygosity (Ho), observed allele number (Na), effective allele number (Ne), marker index (MI), and resolving power (Rp) were observed at 0.63, 2.44, 1.00, 0.60, 0.69, 3.73, 2.60, 1.78 and 4.18, respectively. The minimum and maximum similarity values based on the Jaccard coefficient were observed to be 0.41 to 0.80. The population structure showed an admixture of three different genetic pools in the examined genotypes. The DARwin-based neighbor joining analysis also revealed two genotypes, namely UHFSCr-114 and UHFSCr-122, were more prominent than the rest of the chrysanthemum genotypes. Furthermore, it has already been observed in our previous studies that these two newly evolved genotypes were also found diversified in terms of their phenotypic characteristics. Therefore, in the present study, we observed high genetic variability among the studied genotypes as well as verified the mutant behaviour of newly evolved chrysanthemum genotypes at the molecular level. In addition, the current results would also accelerate and facilitate work on the release of highly diversified chrysanthemum genotypes as new cultivars in the near future.






Similar content being viewed by others
References
Agarwal A, Gupta V, Ul Haq S, Jatav PK, Kothari SL, Kachhwaha S (2019) Assessment of genetic diversity in 29 rose germplasms using SCoT marker. J King Saud Univ Sci 31:780–788. https://doi.org/10.1016/j.jksus.2018.04.022
Anderson NO (2006) Chrysanthemum Dendranthema grandiflora Tzvelv. In: Flower breeding and genetics: issues, challenges, and opportunities for the 21st century. In: Anderson NO (eds). Springer, Dordrecht, pp 389–437
Anonymous (2019) Area and production of horticulture crops for 2018–2019. http://nhb.gov.in/Statistics/Default.aspx
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Short protocols in molecular biology. Wiley, New York, p 836
Caetano-Anolles G, Bassam BJ, Gresshoff PM (1991) DNA fingerprinting using very short arbitrary oligonucleotide primers. Biotechnology 9:553–557. https://doi.org/10.1038/nbt0691-553
Chang L, Dongliang C, Cheng X, Hua L, Yahui LI, Hunag C (2018) SSR analysis of genetic relationship and classification in chrysanthemum germplasm collection. Hortic Plant J 4:73–82. https://doi.org/10.1016/j.hpj.2018.01.003
Datta SKD (2013) Chrysanthemum morifolium (Ramat) a unique genetic material for breeding. Sci Cult 79:307–313
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15. https://doi.org/10.1007/978-3-642-83962-7_18
Earl D, Von Holdt B (2012) Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Feng S, Renfeng H, Jiangjie L, Jiang M, Shen X, Jiang Y, Wang Z, Wang H (2016) Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front Genet 7:13. https://doi.org/10.3389/fgene.2016.00113
Han Z, Ma X, Wei M, Zhao ZR, Chen W (2018) SSR marker development and intraspecific genetic divergence exploration of Chrysanthemum indicum based on transcriptome analysis. BMC Genom 19:1–10. https://doi.org/10.1186/s12864-018-4702-1
Khaing AA, Moe KT, Hong WJ, Park CS, Yeon KH, Park HS, Kim DC, Choi BJ, Jung YJ, Chae SC, Lee KM, Park YJ (2013) Phylogenetic relationships of chrysanthemums in Korea based on novel SSR markers. Genet Mol Res 12:5335–5347. https://doi.org/10.4238/2013.November.7.8
Klie M, Schie S, Linde M, Debener T (2014) The type of ploidy of chrysanthemum is not black or white: a comparison of a molecular approach to published cytological methods. Front Plant Sci 5:479. https://doi.org/10.3389/fpls.2014.00479
Kobeissi B, Saidi A, Kobeissi A, Shafie M (2018) Applicability of SCoT and SSR molecular markers for genetic diversity analysis in Chrysanthemum morifolium genotypes. Proceed National Acad Sci India Sect b Biol Sci 89:1067–1077. https://doi.org/10.1007/s40011-018-1024-7
Kumari S, Choudhary RM, Kumara SRV, Saharan V, Joshi A, Munot M (2017) Assessment of genetic diversity in safflower (Carthamus tinctorius L.) genotypes through morphological and SSR marker. J Pharmacogn Phytochem 6:2723–2731
Li YH, Luo C, Wu ZY, Zhang XH, Cheng X, Dong R, Huang CL (2013) Microsatellite enrichment by magnetic beads in chrysanthemum. Acta Hort 977:269–278. https://doi.org/10.17660/ActaHortic.2013.977.32
Liu H, Zhang QX, Sun M, Pan HT, Kong ZX (2015) Development of expressed sequence tag-simple sequence repeat markers for Chrysanthemum morifolium and closely related species. Genet Mol Res 14:7578–7586. https://doi.org/10.4238/2015.July.13.1
Mekapoku M, Kwon OK, Hyun DY, Lee KJ, Ahn MS, Park JT, Jung JA (2020) Identification of standard type cultivars in chrysanthemum (Dendranthema grandiforum) using SSR markers. Hortic Environ Biotechnol 61:153–161. https://doi.org/10.1007/s13580-019-00201-0
Min-Jo K, Chu H, Lian S, Cho WK (2015) Development of EST-derived SSR markers using next-generation sequencing to reveal the genetic diversity of 50 chrysanthemum cultivars. Biochem Syst Ecol 60:37–45. https://doi.org/10.1016/j.bse.2015.03.002
Negi R, Dhiman SR, Gupta YC (2018) Studies on growth and flowering behavior of newly evolved genotypes of chrysanthemum (Dendranthema grandifloraT zvelev) for cut flower production. Int J Sci Res 8:745–747
Negi R, Dhiman SR, Gupta YC (2019) Studies on growth and flowering behavior of newly evolved genotypes of chrysanthemum (Dendranthema grandiflora Tzvelev) for loose flower production. Int J Curr Microbiol App Sci 8:341–346. https://doi.org/10.20546/ijcmas.2019.811.043
Negi S, Sharma G, Sharma R (2020a) Introgression and confirmation of everbearing trait in strawberry (Fragaria x ananassa Duch.). Physiol Mol Biol Plants 26:2407–2416. https://doi.org/10.1007/s12298-020-00916-w
Negi R, Dhiman SR, Dhiman MR, Gupta YC, Dogra RK, Gupta RK (2020b) Performance and flower characterization of newly evolved genotypes of chrysanthemum (Dendranthema grandiflora Tzvelev) for cut flower production. Int J Chem Stud 8:2921–2923. https://doi.org/10.22271/chemi.2020.v8.i4ai.10093
Negi R, Dogra RK, Dhiman SR, Gupta YC, Gupta RK, Dhiman MR (2020c) Stability analysis of newly evolved genotypes of chrysanthemum (Dendranthema grandiflora Tzvelev) for cut flower production using Eberhart and Russel model. Int J Curr Microbiol Appl Sci 9:477–486. https://doi.org/10.20546/ijcmas.2020c.906.062
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590. https://doi.org/10.1093/genetics/89.3.583
Olejnik A, Parkitna K, Kozak B, Florczak S, Matkowski J, Nowosad K (2021) Assessment of the genetic diversity of chrysanthemum cultivars using SSR markers. Agron 11:2318. https://doi.org/10.3390/agronomy11112318
Park SK, Arens P, Esselink D, Lim JH, Shin HK (2015) Analysis of inheritance mode in chrysanthemum using EST-derived SSR markers. Scientia Hort 192:80–88. https://doi.org/10.1016/j.scienta.2015.05.009
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Roein Z, Asil MH, Sabouri A, Dadras AR (2014) Genetic structure of chrysanthemum genotypes from Iran assessed by AFLP markers and phenotypic traits. Plant Syst Evol 300:493–503. https://doi.org/10.1007/s00606-013-0898-3
Samarina LS, Malyarovskaya VI, Reim S, Yakushina LG, Koninskaya NG, Klemeshova KV, Shkhalakhova RM, Matskiv AO, Shurkina ES, Gabueva TY, Slepchenko NA (2021) Transferability of ISSR, SCoT and SSR markers for Chrysanthemum× Morifolium Ramat and genetic relationships among commercial Russian cultivars. Plants 10:1302. https://doi.org/10.3390/plants10071302
Schlotter C (2004) The evolution of molecular markers- a matter of fashion. Nat Rev Genet 5:63–69. https://doi.org/10.1038/nrg1249
Wang H, Jiang J, Chen S, Qi X, Peng H, Li P, Song A, Guan Z, Fang W, Liao Y, Chen F (2013) Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS ONE 8:e62293. https://doi.org/10.1371/journal.pone.0062293
Yang W, Glover BJ, Rao GY, Yang J (2006) Molecular evidence for multiple polyploidization and lineage recombination in the Chrysanthemum indicum polyploid complex (Asteraceae). New Phytol 171:875–886. https://doi.org/10.1111/j.1469-8137.2006.01779.x
Yeh FC, Yang RC, Boyle T, Ye ZH, Mao JX (1999) Popgene. Microsoft windows-based freeware for population genetic analysis. Release 1:31. http://www.ualberta.ca/~fyeh/,1999-ci.nii.ac.jp
Yuan WJ, Ye S, Du LH, Li SM, Miao X, Shang F (2016) Isolation and characterization of microsatellite markers for Dendranthema morifolium (Asteraceae) using next- generation sequencing. Genet Mol Res 15:1–7. https://doi.org/10.4238/gmr.15048765
Zhang F, Chen S, Chen F, Fang W, Li F (2010) A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Scientia Hort 125:422–428. https://doi.org/10.1016/j.scienta.2010.03.028
Zhang F, Chen S, Chen F, Fang W, Chen Y, Li F (2011) SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol Breed 27:11–23. https://doi.org/10.1007/s11032-010-9409-1
Zhang Y, Dai S, Hong Y, Song X (2014) Application of genomic SSR locus polymorphisms on the identification and classification of chrysanthemum cultivars in China. PLoS ONE 9:e104856. https://doi.org/10.1371/journal.pone.0104856
Zhang Y, Wang C (2013) Assessing the genetic diversity of chrysanthemum cultivars with microsatellites. J Amer Soc Hort Sci 138:479–486. https://doi.org/10.21273/JASHS.138.6.479
Zhao WG, Chung JW, Cho YI, Rha WH, Lee GA, Ma KH, Han SH, Bang KH, Park CB, Kim SM, Park YJ (2010) Molecular genetic diversity and population structure in Lycium accessions using SSR markers. CR Biologies 333:793–800. https://doi.org/10.1016/j.crvi.2010.10.002
Acknowledgements
The authors are grateful to the Indian Council of Agricultural Research (ICAR), New Delhi, India for providing funds through a central assistance scheme.
Author information
Authors and Affiliations
Contributions
AT: conducted PCR-based studies, data recording, writing rough draft of the manuscript; RS: conceptualization, methodology, overall supervision, manuscript editing and finalization; SRD: procurement of genotypes, data recording on phenotypic attributes, manuscript editing; RN: data recording on phenotypic attributes, data analysis; AS: molecular data recording. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Thakur, A., Sharma, R., Dhiman, S.R. et al. Genetic diversity analysis in chrysanthemum (Dendranthema grandiflora Tzvelev) using SSR markers: corroborating mutant behaviour of newly evolved genotypes. Genet Resour Crop Evol 70, 449–460 (2023). https://doi.org/10.1007/s10722-022-01438-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01438-y