Abstract
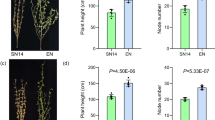
Branch number on the main stem (BNMS) is an important factor that affects crop plant architecture and yield in soybean [Glycine max (L.) Merr.]. With the aim of elucidating the genetic basis of BNMS, 10 consensus quantitative trait loci (QTLs) were identified, which were on 9 chromosomes and explained 0.3–33.3% of the phenotypic variance. One new QTL-qBN.N (R2 = 19.6%) was detected in two populations (F2:3-Taiyuan 2017 and F2:4-Taiyuan 2018), which occur only in Taiyuan. Thus, the interaction analysis of QTL × environment confirmed that QTL-qBN.N was greatly affected by the environment. Of these, QTL-qBN.C2 (R2 = 33.3%) was defined as a major QTL, and was also verified and fine-mapped in the recombinant inbred line population. The high-coverage re-sequencing of two parental lines and newly developed InDel PCR-based markers allowed the region of qBN.C2 to be narrowed down to 304.9 kb. According to the gene annotation of the QTL interval, a total of 24 genes were obtained. Three potential candidate genes were identified by quantitative real-time PCR, and only Glyma.06G188400 had the highest relative expression at the axillary meristem, and thus it may potentially participate in branching development. The successful and rapid fine-mapping and discovery of candidate genes for BNMS in soybean were achieved in this study.




Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotech 30(2): 174–178. https://doi.org/10.1038/nbt.2095
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20(14): 2324–2326. https://doi.org/10.1093/bioinformatics/bth230
Basu D, Tian L, Wang W, Bobbs S, Herock H, Travers A, Showalter AM (2015) A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol 15:295. https://doi.org/10.1186/s12870-015-0670-7
Chen C, Liu M, Jiang L, Liu X, Zhao J, Yan S, Yang S, Ren H, Liu R, Zhang X (2014) Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J Exper Bot 65(17): 4943. https://doi.org/10.1093/jxb/eru258
Chen QS, Zhang ZC, Liu CY, Xin DW, Shan DP, Qiu HM, Shan CY (2007) QTL Analysis of major agronomic traits in soybean. Scientia Agricultura Sinica.
Cheng L (2008) Construction of genetic linkage map and QTL mapping of important traits in soybean [Glycine max (L.) Merrill]. Nanjing Agricultural University
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138(3):963–971
Cline MG (2010) The role of hormones in apical dominance. New approaches to an old problem in plant development. Physiologia Plantarum 90(1): 230–237. https://doi.org/10.1111/j.1399-3054.1994.tb02216.x
Cobb JN, Declerck G, Greenberg A, Clark R, McCouch S (2013) Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. TAG 126(4):867–887. https://doi.org/10.1007/s00122-013-2066-0
Devran Z, Kahveci E, Özkaynak E, Studholme DJ, Tör M (2015) Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing. Mol Breed 35(4):101. https://doi.org/10.1007/s11032-015-0294-5
Ding H (2009) The natural selection effect on resistance to soybean cyst nematode resistance and agronomic traits was distinguished by SSR markers. Nanjing Agricultural University.
Dinka SJ, Campbell MA, Demers T, Raizada MN (2007) Predicting the size of the progeny mapping population required to positionally clone a gene. Genetics 176(4):2035–2054. https://doi.org/10.1534/genetics.107.074377
Doyle JJ (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem bull 19:11–15
Durbin R (2010) Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26(5):589–595. https://doi.org/10.1093/bioinformatics/btp698
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155(1):463
Guan RX. (2004) QTL mapping of soybean agronomic characters and genetic diversityanalysis of soybean cultivara from china and japan. Doctoral dissertation. Chinese Academy of Agricultural Science.
Hayes PM, Liu BH, Knapp SJ, Chen F, Jones B, Blake T, Franckowiak J, Rasmusson D, Sorrells M, Ullrich SE, Wesenberg D, Kleinhofs A (1993) Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. TAG 87(3):392–401. https://doi.org/10.1007/BF01184929
He R, Guan RX, Liu ZX, Zhu XL, Chang RZ, Qiu LQ (2009) Mapping the qBN-cl-l Locus to LG Cl for soybean branching using residual heterozygous lines derived from a segregation population. ScientiaAgriSinica 42(4):1152–1157
Huang ZW, Zhao TJ, Yu DY, Chen SY, Gai JY (2009) Detection of QTLs of yield related traits in soybean. ScientiaAgriSinica 42(12):4155–4165
Huyghe C (1998) Genetics and genetic modifications of plant architecture in grain legumes: a review. Agronomie 18:5–6. https://doi.org/10.1051/agro:19980505
Jiang CZ, Pei CJ, Jing HX, Zhang MC, Wang T, Di R, Liu BQ, Yan L (2011) QTL Analysis of soybean quality and yield related characters. ActaAgriBoreali-Sinica 26(5):127–130
Li W, Zheng DH (2008) QTL mapping for major agronomic traits across two years in soybean (Glycine max L. Merr.). Jcrop Scibiotechnol
Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD, Stacey G (2010) Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol 152(2):541–552. https://doi.org/10.1104/pp.109.148379
Liu B, Wang Y, Zhai W, Deng J, Wang HE, Cui Y, Cheng F, Wang XW, Wu J (2013) Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. TAG 126(1):231–239. https://doi.org/10.1007/s00122-012-1976-6
Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, Hill J, Baum TJ, Cianzio S, Whitham SA, Korkin D, Mitchum MG, Meksem K (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492(7428):256–260. https://doi.org/10.1038/nature11651
Long Y, Shi JQ, Qiu D, Li R, Zhang C, Wang J, Hou J, Zhao J, Shi L, Park BS, Choi SR, Lim YP, Meng JL (2007) Flowering time quantitative trait Loci analysis of oilseed brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177(4):2433–2444. https://doi.org/10.1534/genetics.107.080705
Luo Z, Wang M, Long Y, Huang Y, Shi L, Zhang C, Liu X, Fitt B, Xiang J, Mason AS, Snowdon RJ, Liu P, Meng JL, Zou J (2017) Incorporating pleiotropic quantitative trait loci in dissection of complex traits: seed yield in rapeseed as an example. TAG 130(8):1569–1585. https://doi.org/10.1007/s00122-017-2911-7
Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18(3):586–97. https://doi.org/10.1105/tpc.105.038745
Munier-Jolain NM, Ney B, Duthion C (1996) Analysis of Branching in Spring-sown White Lupins (Lupinus albus L.): The Significance of the Number of Axillary Buds. Annal Bot 77 (2):123–131. https://doi.org/10.1006/anbo.1996.0014
Nelson S (1996) Broadway and the beast: disney comes to times square. TDR 39(2):71
Norsworthy JK, Shipe ER (2005) Effect of row spacing and soybean genotype on mainstem and branch yield. Agron J 97(3):919–923. https://doi.org/10.2134/agronj2004.0271
Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD, Lincoln SE, Lander ES, Tanksley SD (1991) Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127(1):181–197
Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CA, McMullen MD, Holland JB, Bradbury PJ, Buckler ES (2014) The genetic architecture of maize height. Genetics 196(4):1337–1356. https://doi.org/10.1534/genetics.113.159152
Qi X, Li MW, Xie M, Liu X, Ni M, Shao G, Song C, Kay-Yuen Yim A, Tao Y, Wong FL, Isobe S, Wong CF, Wong KS, Xu C, Li C, Wang Y, Guan R, Sun F, Fan G, Xiao Z, Lam HM (2014) Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun 9(5):4340. https://doi.org/10.1038/ncomms5340
Rigsby B, Board JE (2003) Identification of soybean cultivars that yield well at low plant populations. (Crop Ecology, Management & Quality). Crop Sci 43 (1): 234–239. https://doi.org/10.2135/cropsci2003.0234
Sarlikioti V, de Visser PH, Buck-Sorlin GH, Marcelis LF (2011) How plant architecture affects light absorption and photosynthesis in tomato: towards an ideotype for plant architecture using a functional-structural plant model. Annal Bot 108(6):1065–1073. https://doi.org/10.1093/aob/mcr221
Sayama T, Taeyoung H, Yamazaki H, Yamaguchi N, Komatsu K, Takahashi M, Suzuki C, Miyoshi T, Tanaka Y, Xia ZJ, Tsubokura Y, Watanabe S, Harada K, Funatsuki H, Ishimoto M (2010) Mapping and comparison of quantitative trait loci for soybean branching phenotype in two locations. Breed Sci 60(4):380–389. https://doi.org/10.1270/jsbbs.60.380
Schmitz G, Theres K (2005) Shoot and inflorescence branching. Current Opinion Plant Bio 8(5):506–511. https://doi.org/10.1016/j.pbi.2005.07.010
Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. PNAS 99(2):1064–9. https://doi.org/10.1073/pnas.022516199
Shi JQ, Huang SM, Zhan JP, Yu JY, Wang XF, Hua W, Liu SY, Liu GH, Wang HZ (2014) Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res 21(1):53–68. https://doi.org/10.1093/dnares/dst040
Shi JQ, Li RY, Qiu D, Jiang CC, Long Y, Morgan Colin, Bancroft Ian, Zhao JY, Meng JL (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in brassica napus. Genetics 182(3):851–861. https://doi.org/10.1534/genetics.109.101642
Shim S, Kim MY, Ha J, Lee YH, Lee SH (2017) Identification of QTLs for branching in soybean (Glycine max (L.) Merrill). Euphytica 213(9): 225. https://doi.org/10.3390/ijms20010135
Song X, Wei H, Cheng W, Yang S, Zhao Y, Li X,Luo D,Zhang H,Feng X (2015) Development of INDEL markers for genetic mapping based on whole genome resequencing in soybean. G3 5(12): 2793–2799. https://doi.org/10.1534/g3.115.022780
Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES (1992) Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132(3):823–839
Tanksley SD (1993) Mapping polygenes. Annu Rev Genet 27(1):205–233
Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, Ferlanti E, Miller J, Town C, Stuerzlinger W, Provart NJ (2017) ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29(8):1806–1821. https://doi.org/10.1105/tpc.17.00073
Wang J, Zhang X, Lin ZW (2018) QTL mapping in a maize F2 population using Genotyping-by-Sequencing and a modified fine-mapping strategy. Plant Sci 276:171–180. https://doi.org/10.1016/j.plantsci.2018.08.019
Wang Z (2004) Construction of soybean SSR based map and QTL analysis important agronomic traits. GuangXi University
Wang XZ (2008) Inheritance, stability analysis and QTL mapping of yield related traits in soybean. Chinese academy of agricultural sciences
Wei YL (2011) Genetic model analysis and QTL mapping of agronomic and quality traita in aoybean. Henan Agricultural University.
Xu X, Zeng L, Tao Y, Vuong T, Wan J, Boerma R, Noe J, Li Z, Finnerty S, Pathan SM, Shannon JG, Nguyen HT (2013) Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. PNAS 110(33):13469–13474. https://doi.org/10.1073/pnas.1222368110
Yang H, Xue Q, Zhang ZZ, Du JY, Yu DY, Huang F (2018) GmMYB181, a Soybean R2R3-MYB protein, increases branch number in transgenic arabidopsis. Frontiers Plant Sci 18(3):586–597. https://doi.org/10.3389/fpls.2018.01027
Yang ZL, Li GQ (2010) The QTL Analysis of important agronomic traits on a RIL population from a cross between Jinda52 and Jinda57. Acta Agri Boreali-Sinica 25(2):88–92. https://doi.org/10.1111/j.1747-0285.2010.01008.x
Yao D, Liu ZZ, Zhang J, Liu SY, Qu J, Guan SY, Pan LD, Wang D, Liu JW, Wang PW (2015) Analysis of quantitative trait loci for main plant traits in soybean. Genet Mol Res 14(2):6101–6109. https://doi.org/10.4238/2015.June.8.8
Yuan DH (2010) Genetic mechanisms and breeding value analysis of soybean cyst nematode resistance gene. Henan Agricultural University
Zhou R, Wang X, Chen HF, Zhang XJ, Shan ZH, Wu XJ, Cai SP, Qiu DZ, Zhou XA, Wu JS (2009) QTL analysis of lodging and related traits in soybean. Acta Agronomica Sinica 35(1):57–65. https://doi.org/10.3724/SP.J.1006.2009.00057
Funding
This research was supported by the National Key Research and Development Program of China (2016YFD0101500, 2016YFD0101504), Key R & D projects in Shanxi Province (201903D211003), Shanxi Provincial Natural Science Youth Fund (201901D211563), Shanxi Academy of Agricultural Science Postdoctoral Fund (YCX2020BH4), Funded by China Postdoctoral Science Foundation, Shanxi Academy of Agricultural Science Doctoral Research Fund, Shanxi Academy of Agricultural Science Biological Breeding Engineering (17YZGC102) and Shanxi Academy of Agricultural Science Technological Innovation (YGJPY1909).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Y., Lei, Y., Bai, Z. et al. Physical mapping and candidate gene prediction of branch number on the main stem in soybean [Glycine max (L.) Merr.]. Genet Resour Crop Evol 68, 2907–2921 (2021). https://doi.org/10.1007/s10722-021-01163-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01163-y