Abstract
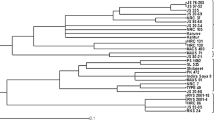
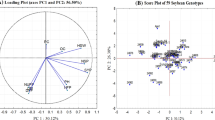
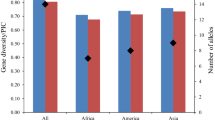
There are numerous soybean [Glycine max (L.) Merr.] breeding programs in Europe focused on development of elite non genetically modified (GM) cultivars for fast growing market of GM-free proteins for animal feed. Due to low variability of visual descriptors and mostly unknown pedigrees, divergent parents’ selection for crosses is a great challenge. Another challenge is cultivar distinction and protection of plant breeders’ rights of ever-increasing number of cultivars. By using 42 microsatellite (SSR) markers, we performed characterization of 97 commercial soybean cultivars and experimental lines developed at various research and breeding institutions in Europe (86) and in North and South America (11) in order to assess their genotype distinction power as well as utility for estimating genetic diversity and genetic structure. A set of 27 most polymorphic SSR markers was sufficient to discriminate all 97 genotypes. Discrimination of, by pedigree very related cultivars, was somewhat difficult due to the low polymorphism but still possible. Cluster analysis showed that European germplasm is mainly distributed into clusters reflecting breeding programs and maturity groups. Performed genetic characterization provides an insight into genetic structure of European soybean germplasm and might serve as a starting point for future breeding decisions. Disclosed SSR data of the analyzed commercial European germplasm can serve for genetic fingerprinting purpose as well as for foundation of public soybean cultivar database.




Similar content being viewed by others
References
Abe J, Xu D, Suzuki Y, Kanazawa A, Shimamoto Y (2003) Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet 106:445–453
Bandillo N, Jarquin D, Song Q, Nelson R, Cregan P, Specht J, Lorenz A (2015) A population structure and genome-wide association analysis on the USDA soybean germplasm collection. The Plant Genome 8:1–13
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Cabezas JA, Ibáñez J, Lijavetzky D, Vélez D, Bravo G, Rodríguez V, Carreño I, Jermakow AM, Carreño J, Ruiz-García L, Thomas MR, Martinez-Zapater JM (2011) A 48 SNP set for grapevine cultivar identification. BMC Plant Biol 11:153
Carter TE Jr, Nelson RL, Sneller CH, Cai Z (2004) Genetic diversity in soybean. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production and uses. Am Soc Agron, Madison, pp 303–406
Cipriani G, Marrazzo MT, Di Gaspero G, Pfeiffer A, Morgante M, Testolin R (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp) genotyping. BMC Plant Biol 8:127
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, van Toai TT, Lohnes DG, Chung J, Specht JE (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
dos Santos JM, Duarte Filho LSC, Soriano ML, da Silva PP, Nascimento VX, de Souza Barbosa GV, Todaroc AR, Netoc CER, Almeida C (2012) Genetic diversity of the main progenitors of sugarcane from the RIDESA germplasm bank using SSR markers. Ind Crop Prod 40:145–150
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor Appl Genet 92:832–839
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
FAOSTAT (2019) Statistics division of the Food and Agriculture Organization (FAO) of the United Nations, Rome. www.fao.org/faostat/en/#data/QC. Accessed 2 June 2019
Gasi F, Kanlić K, Stroil BK, Pojskić N, Asdal Å, Rasmussen M, Kaiser M, Meland M (2016) Redundancies and genetic structure among ex situ apple collections in Norway examined with microsatellite markers. HortScience 51:1458–1462
Gizlice Z, Carter TE Jr, Burton JW (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143–1151
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3.). Department of ecology and evolution, Lausanne University, Switzerland. http://www2.unil.ch/popgen/softwares/fstat.htm. Accessed 7 Nov 2011
Grainger CM, Letarte J, Rajcan I (2018) Using soybean pedigrees to identify genomic selection signatures associated with long-term breeding for cultivar improvement. Can J Plant Sci 98:1176–1187
Gross BL, Volk GM, Richards CM, Forsline PL, Fazio G, Chao CT (2012) Identification of “duplicate” accessions within the USDA-ARS national plant germplasm system Malus collection. J Am Soc Hortic Sci 137:333–342
Guan R, Chang R, Li Y, Wang L, Liu Z, Qiu L (2010) Genetic diversity comparison between Chinese and Japanese soybeans (Glycine max (L.) Merr.) revealed by nuclear SSRs. Genet Resour Crop Evol 57:229–242
Hahn V, Würschum T (2014) Molecular genetic characterization of Central European soybean breeding germplasm. Plant Breed 133:748–755
Hymowitz T (2008) The history of the soybean. In: Johnson LA, White PJ, Galloway R (eds) Soybeans chemistry, production, processing, and utilization. AOCS Press, Urbana, pp 1–31
ISAAA (2017) Pocket K No. 16: Global Status of Commercialized Biotech/GM Crops in 2017. International Service for the Acquisition of Agri-biotech Applications. www.isaaa.org. Retrieved 8 July 2019
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jamali SH, Cockram J, Hickey LT (2019) Insights into deployment of DNA markers in plant cultivar protection and registration. Theor Appl Genet 132:1911–1929
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kurasch AK, Hahn V, Leiser WL, Vollmann J, Schori A, Bétrix CA, Mayr B, Winkler J, Klemens Mechtler K, Aper J, Sudaric A, Pejic I, Sarcevic H, Jeanson P, Balko C, Signor M, Miceli F, Strijk P, Rietman H, Muresanu E, Djordjevic V, Pospišil A, Barion G, Weigold P, Streng S, Krön M, Würschum T (2017) Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant, Cell Environ 40:765–778
Kuroda Y, Tomooka N, Kaga A, Wanigadeva SMSW, Vaughan DA (2009) Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.) and Japanese cultivated soybeans [G. max (L.) Merr.] based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Evol 56:1045–1055
Li YH, Smulders MJ, Chang RZ, Qiu LJ (2011) Genetic diversity and association mapping in a collection of selected Chinese soybean accessions based on SSR marker analysis. Conserv Genet 12:1145–1157
Moreno MV, Nishinakamasu V, Loray MA, Alvarez D, Gieco J, Vicario A, Hopp HE, Heinz RA, Paniego N, Lia VV (2013) Genetic characterization of sunflower breeding resources from Argentina: assessing diversity in key open-pollinated and composite populations. Plant Genet Resources 11:238–249
Oliveira MB, Vieira ESN, Schuster I (2010) Construction of a molecular database for soybean cultivar identification in Brazil. Genet Mol Res 9:705–720
Park SDE (2008) Excel Microsatellite Toolkit. Computer program and documentation distributed by the author. Website http://animalgenomics.ucd.ie/sdepark/ms-toolkit/. Accessed 18 July 2011
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Priolli RHG, Mendes-Junior CT, Arantes NE, Contel EPB (2002) Characterization of Brazilian soybean cultivars using microsatellite markers. Genet Mol Biol 25:185–193
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qiu LJ, Xing LL, Guo Y, Wang J, Jackson SA, Chang RZ (2013) A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol Biol 83:41–50
Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG (2014) STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. SpringerPlus 3:431
Ristova D, Šarčević H, Šimon S, Mihajlov L, Pejić I (2010) Genetic diversity in southeast European soybean germplasm revealed by SSR markers. Agriculturae Conspectus Scientificus 75:21–26
Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosman B, Ganal M (2002) Construction and analysis of a microsatellite-based database of European wheat cultivars. Theor Appl Genet 106:67–73
Signor M (2016) Experiences of soybean cropping and breeding in Northern Italy. Presentation. Meeting Sustainable Soya—Sustainable Europe- Budapest, 24–25 Nov 2016
Sohn HB, Kim SJ, Hwang TY, Park HM, Lee YY, Markkandan K, Lee D, Lee S, Hong SY, Song YH, Koo BC, Kim YH (2017) Barcode system for genetic identification of soybean [Glycine max (L.) Merrill] cultivars using InDel markers specific to dense variation blocks. Front Plant Sci 8:520
Song QJ, Quigley CV, Nelson RL, Carter TE, Boerma HR, Strachan JL, Cregan PB (1999) A selected set of trinucleotide simple sequence repeat markers for soybean cultivar identification. Plant Variet Seeds 12:207–220
Song Q, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB (2015) Fingerprinting soybean germplasm and its utility in genomic research. Genes Genom Genet 5:1999–2006
SoyBase (2018) Soybean breeder’s toolbox. http://soybase.org. Accessed May 2018
Tavaud-Pirra M, Sartre P, Nelson R, Santoni S, Texier N, Roumet P (2009) Genetic diversity in a soybean collection. Crop Sci 49:895–902
This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, Crespan M, Dangl GS, Eisenheld C, Ferreira-Monteiro F, Grando S, Ibáñez J, Lacombe T, Laucou V, Magalhães R, Meredith CP, Milani N, Peterlunger E, Regner F, Zulini L, Maul E (2004) Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet 109:1448–1458
USDA - U.S. Department of Agriculture (2019) USDA Agricultural Projections to 2028. U.S. Department of Agriculture Long-term Projections Report OCE-2019-1. Accessed September 2019
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256
Wang L, Guan R, Zhangxiong L, Chang R, Qiu L (2006) Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci 46:1032–1038
Widaningsih NA, Purwanto E, Nandariyah N, Reflinur R (2014) The use of DNA microsatellite markers for genetic diversity identification of soybean (Glycine max (L) Meriil.) as a supplementary method in reference collections management. Indones J Biotechnol 19:136–145
Wysmierski PT, Vello NA (2013) The genetic base of Brazilian soybean cultivars: evolution over time and breeding implications. Genet Mol Biol 36:547–555
Xiong D, Zhao T, Gai J (2010) Genetic bases of improved soybean cultivars released from 1923 to 2005 in China—a historical review. Front Agric China 4:383–393
Yoon MS, Song QJ, Choi IY, Specht JE, Hyten DL, Cregan PB (2007) BARCSoySNP23: a panel of 23 selected SNPs for soybean cultivar identification. Theor Appl Genet 114:885–899
Acknowledgements
This research was funded by the Environmental Protection and Energy Efficiency Fund with the support of the Croatian Science Foundation of the Republic of Croatia and partly supported by the project KK.01.1.1.01.0005, Centre of Excellence for Biodiversity and Molecular Plant Breeding (CoE CroP-BioDiv), Zagreb, Croatia. Special thanks to Dr. Marco Signor, Regional Agriculture Agency of Friuli Venezia Giulia (ERSA), Udine, Italy, who provided for this research number of genotypes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
List of selected SSR loci (DOCX 30 kb)
Online resource 2
Genetic profiles of 97 soybean genotypes across 42 SSR loci. Allele values are given in base pairs (XLSX 45 kb)
Online resource 3
Range, alleles and major allele frequencies for 42 SSR loci. (DOCX 27 kb)
Rights and permissions
About this article
Cite this article
Žulj Mihaljević, M., Šarčević, H., Lovrić, A. et al. Genetic diversity of European commercial soybean [Glycine max (L.) Merr.] germplasm revealed by SSR markers. Genet Resour Crop Evol 67, 1587–1600 (2020). https://doi.org/10.1007/s10722-020-00934-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-00934-3