Abstract
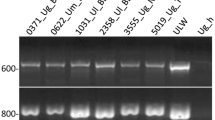
We studied the genetic relationships of Lactuca georgica samples originating in Armenia and the Russian Federation with samples representing four other predominantly self-pollinating wild Lactuca species (L. serriola, L. aculeata, L. saligna, and L. virosa) originating in various countries, as well as with samples representing cultivated lettuce, L. sativa by using 48 TRAP markers. We also visualized their genetic diversity and structure. The present study is likely the first molecular phylogenetic evaluation of a detailed screening of L. georgica germplasm. Data analysis of the three major wild species in this study, L. georgica (134 samples), L. virosa (57 samples), and L. serriola (40 samples) showed that allele frequencies of all 47 polymorphic loci varied significantly among the species. A total of 11, 9, and 10 alleles were unique to L. georgica, L. serriola, and L. virosa, respectively; 71% of TRAP marker diversity was between species. The Neighbor-Joining tree clearly clustered the whole set of 238 samples according to their taxonomic determination. It also reflects the gene diversity as well as the genetic distance values among samples representing the between and within variance of the various species. The L. georgica samples clustered most distantly from the L. sativa samples. The interspecies comparisons between samples belonging to L. georgica with those belonging to L. sativa displayed a high distance, lower only from the interspecies comparisons between samples belonging to L. virosa (in the tertiary gene pool of cultivated lettuce) with those belonging to L. sativa. Thus, additional molecular data with more hybridization experiments are necessary to reconsider if L. georgica is indeed a constituent of the primary gene pool of cultivated lettuce. The L. georgica samples were divided into two sub-clusters, with samples collected in southeast and central Armenia grouping together while all those collected in the north and Dagestan grouped together.



Similar content being viewed by others
References
Aiken HH (1955) Tables of cumulative binomial probability distribution. Harvard University Press, Cambridge
Beharav A, Cahaner A, Pinthus MJ (1994) Mixed model for estimating the effects of the Rht1 dwarfing allele, background genes, CCC and their interaction on culm and leaf elongation of spring wheat (Triticum aestivum L.). Heredity 72:237–241
Beharav A, Stojakowska A, Ben-David R, Malarz J, Michalska K, Kisiel W (2015) Variation of sesquiterpene lactone contents in Lactuca georgica natural populations from Armenia. Genet Resour Crop Evol 62:431–441
Doležalová I, Křístková E, Lebeda A, Vinter V (2002) Description of morphological characters of wild Lactuca L. spp. genetic resources (English–Czech version). Hort Sci 29:56–83
Doležalová I, Křístková E, Lebeda A, Vinter V, Astley D, Boukema IW (2003) Basic morphological descriptors for genetic resources of wild Lactuca spp. Plant Gen Resour Newslett 134:1–9
Gabrielian E, Fragman-Sapir O (2008) Flowers of the Transcaucasus and adjacent areas: including Armenia, Eastern Turkey, Southern Georgia, Azerbaijan and Northern Iran. A.R.G Gantner Verlag KG, Ruggell
Gabrielian E, Zohary D (2004) Wild relatives of food crops native to Armenia and Nakhichevan. Flora Mediterranea, vol 14. Edinburgh University Press, Palermo, pp 5–80
Gedil MA, Slabaugh MB, Berry S, Johnson R, Michelmore R, Miller J, Gulya T, Knapp SJ (2001) Candidate disease resistance genes in sunflower cloned using conserved nucleotide-binding site motifs: genetic mapping and linkage to the downy mildew resistance gene Pl1. Genome 44:205–212
Globerson D, Netzer D, Sacks J (1980) Wild lettuce as a source for improving cultivated lettuce. In: Proceedings Eucarpia meeting on leafy vegetables, Littlehampton, pp 86–96
Hartl DL (1980) Principles of population genetics. Sinauer Associates, Inc., Sunderland
Hu J, Vick B (2003) Target region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol Rep 21:289–294
Hu J, Ochoa OE, Truco MJ, Vick BA (2005) Application of the TRAP technique to lettuce (Lactuca sativa L.) genotyping. Euphytica 144:225–235
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kilian N, Gemeinholzer B, Lack HW (2009) Tribe Cichorieae. In: Funk VA, Susanna A, Stuessy T, Bayer R (eds) Systematics, evolution, and biogeography of the Compositae. International Association for Plant Taxonomy, Vienna, pp 343–383
Kisha TJ, Johnson RC, Skinner DZ, Green SL (2002) Variance of genetic distance measurement among alfalfa accessions compared from several molecular marker type. Crop Science Society of America, Madison, p 175620
Kitner M, Majesky L, Křistková E, Jemelková M, Lebeda A, Beharav A (2015) Genetic structure and diversity in natural populations of three predominantly self-pollinating wild Lactuca species in Israel. Genet Resour Crop Evol 62:991–1008
Kwon SJ, Smŷkal P, Hu J, Wang M, Kim SJ, McGee RJ, McPhee K, Coyne CJ (2013) User-friendly markers linked to Fusarium wilt race 1 resistance Fw gene for marker-assisted selection in pea. Plant Breed 132:642–648
Lebeda A, Doležalová I, Astley D (2004a) Representation of wild Lactuca spp. (Asteraceae, Lactuceae) n world genebank collections. Genet Resour Crop Evol 51:167–174
Lebeda A, Doležalová I, Feráková V, Astley D (2004b) Geographical distribution of wild Lactuca species (Asteraceae, Lactuceae). Bot Rev 70:328–356
Lebeda A, Doležalová I, Novotná A (2012) Wild and weedy Lactuca species, their distribution, ecogeography and ecobiology in USA and Canada. Genet Resour Crop Evol 59:1805–1822
Lebeda A, Křístková E, Doležalová I, Kitner M, Widrlechner MP (2018) Wild Lactuca species in North America, Chapter 15. In: Crop wild relatives and wild utilized plants in North America. Springer (in press)
Lebeda A, Křístková E, Kitner M, Mieslerová B, Jemelková M, Pink DAC (2014) Wild Lactuca species, their genetic diversity, resistance to diseases and pests, and exploitation in lettuce breeding. Eur J Plant Pathol 138:597–640
Lebeda A, Křístková E, Kitner M, Mieslerová B, Pink DA (2016) Wild Lactuca saligna: a rich source of variation for lettuce breeding. In: Maxted N, Ehsan Dulloo M, Ford-Lloyd BV (eds) Enhancing crop genepool use: capturing wild relative and landrace diversity for crop improvement. CAB International, Wallingford, pp 32–46
Lebeda A, Ryder EJ, Grube R, Doležalová I, Křístková E (2007) Lettuce (Asteraceae Lactuca spp.). In: Singh RJ (ed) Genetic resources, chromosome engineering, and crop improvement, vegetable crops, vol 3. CRC Press, Boca Raton, pp 377–472
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Li J, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Lynch M, Milligan B (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Maxted N, Kel SP, Ford-Lloyd BV (2008) Crop wild relative conservation and use: establishing the context. In: Maxted N et al (eds) Crop wild relative conservation and use. CABI, Wallingford, pp 3–30
Michalska K, Beharav A, Kisiel W (2014) Sesquiterpene lactones from roots of Lactuca georgica. Phytochem Lett 10:10–12
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Owuor ED, Beharav A, Fahima T, Kirzhner VM, Korol AB, Nevo E (2003) Microscale ecological stress causes RAPD molecular selection in wild barley, Neve Yaar microsite, Israel. Genet Resour Crop Evol 50:213–224
Rohlf FJ (2009) NTSYSpc: numerical taxonomy system, ver. 2.21i. Exeter Software, Setauket
Suman A, Ali K, Arro J, Parco AS, Kimbeng CA, Baiskh N (2012) Molecular diversity among members of the Saccharum complex assessed using TRAP markers based on lignin-related genes. Bioenergy Res 5:197–205
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
van Treuren R, Coquin P, Lohwasser U (2012) Genetic resources collections of leafy vegetables (lettuce, spinach, chicory, artichoke, asparagus, lamb’s lettuce, rhubarb and rocket salad): composition and gaps. Genet Resour Crop Evol 59:981–997
van Treuren R, van Hintum TJL (2009) Comparison of anonymous and targeted molecular markers for the estimation of genetic diversity in ex situ conserved Lactuca. Theor Appl Genet 119:1265–1279
Wei Z, Zhu S-X, Van den Berg RG, Bakker FT, Schranz ME (2017) Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparison. Genet Resour Crop Evol 64:55–71
Weir BS (1996) Genetic Data Analysis II. Methods for discrete population genetic data. Sinauer Associates, Inc., Sunderland, p 356
Wright S (1946) Isolation by distance under diverse systems of mating. Genetics 31:39–59
Yang TJ, Jang SW, Kim WB (2007) Genetic relationships of Lactuca spp. revealed by RAPD, Inter-SSR, AFLP, and PCR-RFLP analyses. J Crop Sci Biotechnol 10:29–34
Yeh FC, Yang RC, Boyle T (1999) POPGENE version 1.32: microsoft windows-based freeware for population genetic analysis, quick user guide. Center for International Forestry Research, University of Alberta, Edmonton
Zohary D (1991) The wild genetic resources of cultivated lettuce (Lactuca sativa L.). Euphytica 53:31–35
Acknowledgements
AB wishes to thank Dr. Alvina Avagian (National Coordinator for plant genetic resources, Armenia) for excellent organization of germplasm collections; Dr. Margarita Harutyunyan, Mrs. Marina Hovhannisyan, and Mrs. Ani Petrosyan [Laboratory of Plants Gene Pool and Breeding, Armenian National Agrarian University (ANAU), Yerevan, Armenia], and Dr. Gayane Melyan (Gene Bank of Plant Genetic Resources for Food and Agriculture, Scientific Center of Agrobiotechnology of ANAU, Echmiadzin, Armenia) for their great technical assistance during a collection trips in Armenia in 2011. BH wishes to thank Dr. George Fayvush (Institute of Botany, Yerevan, NAS of Armenia), Prof. M. Mosulishvili (Institute of Ecology, Tbilisi, Georgia), Dr. S. Litvinskaya (Kuban State University, Krasnodar, Russian Federation) and Dr. R. Murtazaliev, Mountain Botanical Garden, RAS Dagestan Scientific Center, Makhachkala, Dagestan) for seed collecting in 2009 and 2010. KLR wishes to thank Chris Richards (USDA-ARS, Fort Collins, CO) for his assistance with bootstrap analysis. AL wishes to thank the Internal Granting Agency of Palacký University in Olomouc, Czech Republic (IGA_Prf_2018-001), Project MSM 6198959215 (Ministry of Education, Youth and Sports, Czech Republic). A special thank to Dr. Matěj Lövy (Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic) for his production of Fig. 1. This research was supported by USDA-ARS CRIS Project 5348-21000-026-00D and NIFA MultiState Project W006.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Beharav, A., Hellier, B., Richardson, K.L. et al. Genetic relationships and structured diversity of Lactuca georgica germplasm from Armenia and the Russian Federation among other members of Lactuca L., subsection Lactuca L., assessed by TRAP markers. Genet Resour Crop Evol 65, 1963–1978 (2018). https://doi.org/10.1007/s10722-018-0669-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0669-7