Abstract
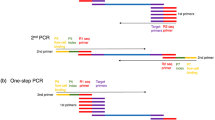
The advent of large-scale DNA sequencing technology has generated a tremendous amount of sequence information for many important organisms. We have developed a rapid and efficient PCR-based technique, which uses bioinformatics tools and expressed sequence tag (EST) database information to generate polymorphic markers around targeted candidate gene sequences. This target region amplification polymorphism (TRAP) technique uses 2 primers of 18 nucleotides to generate markers. One of the primers, the fixed primer, is designed from the targeted EST sequence in the database; the second primer, the arbitrary primer, is an arbitrary sequence with either an AT-or GC-rich core to anneal with an intron or exon, respectively. PCR amplification is run for the first 5 cycles with an annealing temperature of 35°C, followed by 35 cycles with an annealing temperature of 50°C. For different plant species, each PCR reaction can generate as many as 50 scorable fragments with sizes ranging from 50–900 bp when separated on a 6.5% polyacrylamide sequencing gel. The TRAP technique should be useful in genotyping germplasm collections and in tagging genes governing desirable agronomic traits of crop plants.
Similar content being viewed by others
Abbreviations
- EST:
-
expressed sequence tag
- nt:
-
nucleotide
- PCR:
-
polymerase chain reaction
- TRAP:
-
target region amplification polymorphism
References
Compositae Genome Project. UC Davis. 21 June 2003 <http://cgpdb.ucdavis.edu/database/php_my_admin/php_my_admin.php>.
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp.japonica). Science 296: 92–100.
Gupta PK and Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113: 163–185.
Hu J, Seiler GJ, Jan CC, and Vick BA (2003) Assessing genetic variability among sixteen perennialHelianthus species using PCR-based TRAP markers. <http://www. sunflowernsa.com/research_statistics/research_workshop/documents/88.PDF>. Proc. 25th Sunflower Res. Workshop, 16–17 Jan. 2003. Fargo, ND.
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sale F, Van de Wiel C, Bredemeijer G, Buiatti M, Maestri E, Malcevshi A, et al. (1997) Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3: 381–390.
Li G and Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging inBrassica. Theor Appl Genet 103: 455–461.
Meinke DW, Cherry JM, Dean C, Rounsley SD, and Koorneef M (1998)Arabidopsis thaliana: A model plant for genome analysis. Science 282: 662–682.
Michelmore RW, Paran I, and Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genome regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832.
Rozen S and Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S and Misener S (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology, pp, 365–386, Humana Press, Totowa, NJ.
TheArabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plantArabidopsis thaliana. Nature 408: 796–815.
Virk PS, Zhu J, Newbury HJ, Bryan GJ, Jackson MT, and Ford-Lloyd BV (2000) Effectiveness of different classes of molecular markers for classifying and revealing variations in rice (Oryza sativa) germplasm. Euphytica 112: 275–284.
Vos P, Hogers R, Bleeker M, Reijans M, Van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, and Zabeau M (1995) AFLP: a new technique for DNA finger-printing. Nucl Acids Res 23: 4407–4414.
Whitehead Institute for Biomedical Research. 21 June 2003 <http://www-genome.wi.mit. edu/cgi-bin/primer/primer3.cgi>.
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, and Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18: 6231–6235.
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. (2002) A draft sequence of the rice Genome (Oryza sativa L. ssp.indica). Science 296: 79–92.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hu, J., Vick, B.A. Target region amplification polymorphism: A novel marker technique for plant genotyping. Plant Mol Biol Rep 21, 289–294 (2003). https://doi.org/10.1007/BF02772804
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF02772804