Abstract
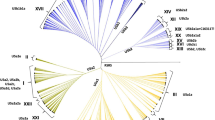
The genus Musa L. comprises of economically important bananas and has been divided into five sections based on their chromosome number and morphological characteristics viz., Australimusa, Callimusa, Eumusa, Rhodochlamys and Ingentimusa. However, this sectional classification has long been disputed. In this study, we present data on sequenced multiple DNA fragments from the maternally inherited chloroplast genome (atpB-rbcL spacer, trnK-matK intron and psbK-psbI spacer) and the biparentally inherited internal transcribed spacers of nuclear ribosomal DNA of three species of Musa along with data available in GenBank database to understand their implications on the sectional relationship and phylogeny of the genus Musa. In our findings, none of the five sections of Musa previously defined based on morphology was recovered in the molecular phylogeny analysis using cpDNA and nrDNA. Instead, the results corroborate with the suggestion that sectional classification in the genus Musa should be reviewed by combining together the section Eumusa and Rhodochlamys as one and section Australimusa and Callimusa as a single different section, along with that of Ingentimusa.



Similar content being viewed by others
References
Andreasen K, Bremer B (2000) Combined phylogenetic analysis in the Rubiaceae–Ixoroideae: morphology, nuclear and chloroplast DNA data. Am J Bot 87:1731–1748
Argent GCG (1976) The wild bananas of Papua New Guinea. Notes on Royal Botanic Garden, vol 35. Edinburgh, pp 77–114
Baker JG (1893) A synopsis of the genera and species of Museae. Ann Bot 7:189–229
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol 1:3–16
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA-A valuable source of evidence on Angiosperm phylogeny. Ann Mo Bot Gard 82:247–277
Bartos J, Alkhimova O, Dolezelova M, De Langhe E, Dolezel J (2005) Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): taxonomic implications. Cytogenet Genome Res 109:50–57
Bekele E, Shigeta M (2011) Phylogenetic relationships between Ensete and Musa species as revealed by the trnT trnF region of cpDNA. Genet Resour Crop Evol 58:259–269
Bhat KV, Bhat SR, Chandel KPS (1992) Survey of isozyme polymorphism for clonal identification in Musa I esterase, acid phosphatase and catalase. J Hortic Sci 67:501–507
Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD et al (1993) Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard 80:528–580
Cheesman EE (1947) Classification of the bananas II. The genus Musa L. Kew Bull 2:106–117
Cheesman EE (1948a) Classification of the bananas III. Critical notes on species. Kew Bull 3(11–28):145–157
Cheesman EE (1948b) Classification of the bananas III. Critical notes on species. Kew Bull 3:323–328
Cheesman EE (1949a) Classification of the bananas III. Critical notes on species. Kew Bull 4:23–28, 133–137, 265–272
Cheesman EE (1949b) Classification of the bananas III.Critical notes on species. Kew Bull 4:445–452
Cheesman EE (1950) Classification of the bananas III. Critical notes on species. Kew Bull 5(27–31):151–155
Christelová P, Valárik M, Hřibová E, De Langhe E, Doležel J (2011) A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evol Biol 11:103. doi:10.1186/1471-2148-11-103
Čížková J, Hřibová E, Christelová P, Van den Houwe I, Häkkinen M, Roux N, Swennen R, Doležel J (2015) Molecular and cytogenetic characterization of wild Musa species. PLoS One 10(8):e0134096. doi:10.1371/journal.pone.0134096
Daniells JW, Jenny C, Karamura DA, Tomekpe K, Arnaud E, Sharrock S (2001) Musalogue: a catalogue of Musa germplasm diversity in the genus Musa. INIBAP, France
De Langhe E (2000) Diversity in the genus Musa, its significance and its potential. Acta Hortic 540:81–86
de Queiroz A, Donoghue MJ, Kim J (1995) Separate versus combined analysis of phylogenetic evidence. Annu Rev Ecol Evol Syst 26:657–681
Gawel NJ, Jarret RL (1991a) A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep 9:262–266
Gawel NJ, Jarret RL (1991b) Chloroplast DNA restriction fragment length polymorphisms (RFLP) in Musa species. Theor Appl Genet 81:783–786
Gawel NJ, Jarret RL, Whittemore AP (1992) Restriction fragment length polymorphism (RFLP)-based phylogenetic analysis of Musa. Theor Appl Genet 84:286–290
Gielly L, Taberlet P (1994) The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. Mol Biol Evol 11:769–777
Godwin ID, Aitken EAB, Smith LW (1997) Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 18:1524–1528
Grapin A, Noyer JL, Carreel F, Dambler D, Baurens FC, Lanaud C, Lagoda PJL (1998) Diploid Musa acuminata genetic diversity assayed with sequence tagged microsatellite sites. Electrophoresis 19:1374–1380
Häkkinen M (2013) Reappraisal of sectional taxonomy in Musa (Musaceae). Taxon 62:809–813
Hoot SB, Culham A, Crane PR (1995) The utility of atpB gene sequences in resolving phylogenetic relationships: comparison with rbcL and 18S ribosomal DNA sequences in the Lardizabalaceae. Ann Mo Bot Gard 82:194–207
Howell EC, Newbury HJ, Swennen RL, Withers LA, Ford-Lloyd BV (1994) The use of RAPD for identifying and classifying Musa germplasm. Genome 37:328–332
Hřibová E, Čížková J, Christelová P, Taudien S, de Langhe E, Doležel J (2011) The ITS1-5.8S-ITS2 sequence region in the Musaceae: structure, diversity and use in molecular phylogeny. PLoS One 6(3):e17863
Huelsenbeck JP, Bull JJ, Clifford WC (1996) Combining data in phylogenetic analysis. Trends Ecol Evol 11:152–158
INIBAP/CIRAD (1996) Descriptors for Banana (Musa spp.). INIBAB, Montpellier
Jarret RL, Gawel NJ (1995) Molecular markers, genetic diversity and systematic. In: Gowen S (ed) Bananas and plantains. Chapman and Hall, London, pp 67–83
Johnson LA, Soltis DE (1994) MatK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Syst Bot 19:143–156
Jussieu AL (1789) Ordo I. Musae Genera plantarum: secundum ordines naturals disposita, juxta methodum in Horto regio parisiensi exaratam, anno MDCCLXXIV, Paris, pp 61–62
Källersjö M, Farris JS, Chase MW, Bremer B, Fay MF, Humphries CJ et al (1998) Simultaneous parsimony jackknife analysis of 2538 rbcL DNA sequences reveals support for major clades of green plants, land plants, seed plants, and flowering plants. Plant Sys Evol 213:259–287
Karamura DA (1999) Numerical taxonomic studies of the East African Highland bananas (Musa AAA-East Africa) in Uganda. PhD thesis from the University of Reading published by INIBAP, Montpellier, France
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261
Kress WJ, Specht CD (2005) Between cancer and capricorn: phylogeny, evolution, and ecology of the tropical Zingiberales. In: Friis I, Balslev H (eds) Plant diversity and complexity patterns—local, regional and global dimensions, vol 55., Biologiske SkrifterThe Royal Danish Academy of Sciences and Letters, Copenhagen, pp 459–478
Kress WJ, Prince LM, Hahn WJ, Zimmer EA (2001) Unraveling the evolutionary radiation of the families of the Zingiberales using morphological and molecular evidence. Syst Biol 50:926–944
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G et al (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105:2923–2928
Lanaud C, Tezenas du Montcel H, Jolivot MP, Glaszmann JC, Gonzalez de Leon D (1992) Variation in ribosomal gene spacer length among wild and cultivated bananas. Heredity 68:148–156
Li LF, Hakkinen M, Yuan YM, Hao G, Ge XJ (2010) Molecular phylogeny and systematics of the banana family (Musaceae) inferred from multiple nuclear and chloroplast DNA fragments, with a special reference to the genus Musa. Mol Phylogenet Evol 57:1–10
Linnaeus C (1753) Musa L. species plantarum, vol 2. Impensis Laurentii Salvii, Stockholm, p 1043
Liu AZ, Kress J, Li DZ (2010) Phylogenetic analyses of the banana family (Musaceae) based on nuclear ribosomal (ITS) and chloroplast (trnL-F) evidence. Taxon 59:20–28
Nixon KC, Carpenter JM (1997) On simultaneous analysis. Cladistics 12:221–241
Nwakanma DC, Pillay M, Okoli BE, Tenkouano A (2003) Sectional relationships in the genus Musa L. inferred from the PCR-RFLP of organelle DNA sequences. Theor Appl Genet 107:850–856
Osuji JO, Crouch J, Harrison G, Heslop-Harrison JS (1997) Identification of the genomic constitution of Musa L lines (bananas, plantains and hybrids) using molecular cytogenetics. Ann Bot 80:787–793
Purseglove JW (1972) Tropical crops. Monocotyledons, vol 2. Longmans, London
Qiu YL, Lee JH, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Al ET (1999) The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402:404–407
Rieseberg LH, Wendel JF (1993) Introgression and its consequences in plants. In: Harrison R (ed) Hybrid zones and the evolutionary process. Oxford University Press, Oxford, pp 70–109
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sagot P (1887) Sur le genre bananier. B Soc Bot Fr 34:328–330
Shepherd K (1990) Observations on Musa taxonomy. In: Jarret RL (ed) Identification of genetic diversity in the genus Musa: Proceedings of an international workshop held at Los Banos, Philippines, 5–10 September 1988 INIBAP, Montpellier, pp 158–165
Shepherd K (1999) Cytogenetics of the genus Musa. International network for the improvement of banana and plantain, Montpellier
Simmonds NW (1954) Isolation in Musa, sections Eumusa and Rhodochlamys. Evolution 8:65–74
Simmonds NW (1962) The evolution of the bananas. Longmans, London
Simmonds NW, Weatherup STC (1990) Numerical taxonomy of the wild bananas (Musa). New Phytol 115:567–571
Small RL, Lickey EB, Shaw J, Hauk WD (2005) Amplification of noncoding chloroplast DNA for phylogenetic studies in lycophytes and monilophytes with a comparative example of relative phylogenetic utility from Ophioglossaceae. Mol Phylogenet Evol 36:509–522
Soltis DE, Soltis PS (1998) Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematics of plants II-DNA Sequencing. Kluwer Academic Publishers, Boston, pp 1–42
Soltis PS, Soltis DE, Chase MW (1999) Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402:402–404
Stover RH, Simmonds NW (1987) Bananas. Longman Scientific and Technical, Essex
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for the amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson J, Higgins D, Gibson T (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4690
Tsumura Y, Kawahara T, Wickneswari R, Yoshimura K (1996) Molecular phylogeny of Dipterocarpaceae in Southeast Asia using RFLP and PCR-amplified chloroplast genes. Theor Appl Genet 93:22–29
Ude G, Pillay M, Nwakanma D, Tenkouano A (2002) Analysing of genetic diversity and sectional relationships in Musa using AFLP markers. Theor Appl Genet 104:1239–1245
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Wong C, Kiew R, Argent G, Set O, Lee SK, Gan YY (2002) Assessment of the validity of the sections in Musa (Musaceae) using AFLP. Ann Bot 90:231–238
Wong C, Argent GCG, Kiew R, Set O, Gan YY (2003) The genetic relations of Musa species from Mount Jaya, New Guinea, and reappraisal of the sections of Musa (Musaceae). Gard Bull Singapore 55:97–111
Xu F, Sun M (2001) Comparative analysis of phylogenetic relationships of grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribed spacer, amplified fragment length polymorphism, and double-primer fluorescent intersimple sequence repeat markers. Mol Phylogenet Evol 21:372–387
Acknowledgments
We are thankful to the Head, Department of Biotechnology and Bioinformatics, North-Eastern Hill University for providing necessary facilities to carry out this research work. Immense appreciation are also accorded to Dr. Jeremy Dkhar, DST-INSPIRE faculty, Jawaharlal Nehru University, New Delhi and all the Plant Biotechnology Laboratory members, North-Eastern Hill University for all the assistance and motivation rendered.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lamare, A., Otaghvari, A.M. & Rao, S.R. Phylogenetic implications of the internal transcribed spacers of nrDNA and chloroplast DNA fragments of Musa in deciphering the ambiguities related to the sectional classification of the genus. Genet Resour Crop Evol 64, 1241–1251 (2017). https://doi.org/10.1007/s10722-016-0433-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0433-9