Abstract
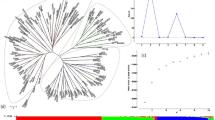
Colored barley is used for the development of barley-containing functional food because of the special nutrients present in its grains. Meanwhile, grain color, which is also one of the primary traits selected in crop domestication, can be used as an important indicator of crop evolution and domestication. In this study, 35 SSR marker pairs were used to analyze the genetic diversity of 277 germplasm accessions representing yellow, purple, blue, and black barley. The analysis revealed that rich genetic variation was mainly caused by the genetic diversity of individuals. Differently colored barley populations could be further divided into subpopulations. As per PCoA, AMOVA, and structure analysis, a complex evolutionary relationship existed among the four colored barley populations. This phenomenon may have been the result of long-term multidirectional selection by humans on the basis of barley use. Studies on colored barley provide useful data for future breeding and utilization of these materials.



Similar content being viewed by others
References
Abdel-Aal E-SM, Young JC, Rabalski I (2006) Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J Agric Food Chem 54:4696–4704
Badr A, Sch R, El Rabey H, Effgen S, Ibrahim H, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bellido GG, Beta T (2009) Anthocyanin composition and oxygen radical scavenging capacity (ORAC) of milled and pearled purple, black, and common barley. J Agric Food Chem 57:1022–1028
Blattner FR, Méndez AGB (2001) RAPD data do not support a second centre of barley domestication in Morocco. Genet Resour Crop Evol 48:13–19
Chen X, Guo S, Chen D, Liu P, Jia X, Sun L (2006) Assessing genetic diversity of Chinese cultivated barley by STS markers. Genet Resour Crop Evol 53:1665–1673
Chen Z, Lu R, Zou L, Du Z, Gao R, He T, Huang J (2012) Genetic diversity analysis of barley landraces and cultivars in the Shanghai region of China. Genet Mol Res 11:644–650
Choo TM, Vigier B, Ho KM, Ceccarelli S, Grando S, Franckowiak JD (2005) Comparison of black, purple, and yellow barleys. Genet Resour Crop Evol 52:121–126
Dai F, Nevo E, Wu D, Comadran J, Zhou M, Qiu L, Chen Z, Beiles A, Chen G, Zhang G (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci 109:16969–16973
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Feng Z-Y, Zhang L-L, Zhang Y-Z, Ling H-Q (2006) Genetic diversity and geographical differentiation of cultivated six-rowed naked barley landraces from the Qinghai-Tibet plateau of China detected by SSR analysis. Genet Mol Biol 29:330–338
Gong X, Scoles G, Westcott S, Li C, Yan G, Lance R, Sun D (2009) Comparative analysis of genetic diversity between Qinghai-Tibetan wild and Chinese landrace barley. Genome 52:849–861
Hamza S, Hamida WB, Rebaï A, Harrabi M (2004) SSR-based genetic diversity assessment among Tunisian winter barley and relationship with morphological traits. Euphytica 135:107–118
Hou Y, Yan Z, Wei Y, Zheng Y (2005) Genetic diversity in barley from west China based on RAPD and ISSR analysis. Barley Genet Newsl 35:9–22
Kim M-J, Hyun J-N, Kim J-A, Park J-C, Kim M-Y, Kim J-G, Lee S-J, Chun S-C, Chung I-M (2007) Relationship between phenolic compounds, anthocyanins content and antioxidant activity in colored barley germplasm. J Agric Food Chem 55:4802–4809
Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L, Lin F, Luan W, Chen P, Yan Z (2008) Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet 117:857–871
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Lundqvist U, Franckowiak J, Konishi T (1997) New and revised descriptions of barley genes. Barley Genet Newsl 26:4–8
Matus I, Hayes P (2002) Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. Genome 45:1095–1106
Mayer K, Waugh R, Brown J, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716
Morrell PL, Clegg MT (2007) Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci 104:3289–3294
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Orabi J, Backes G, Wolday A, Yahyaoui A, Jahoor A (2007) The Horn of Africa as a centre of barley diversification and a potential domestication site. Theor Appl Genet 114:1117–1127
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petersen L, Østergård H, Giese H (1994) Genetic diversity among wild and cultivated barley as revealed by RFLP. Theor Appl Genet 89:676–681
Pillen K, Binder A, Kreuzkam B, Ramsay L, Waugh R, Förster J, Leon J (2000) Mapping new EMBL-derived barley microsatellites and their use in differentiating German barley cultivars. Theor Appl Genet 101:652–660
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Russell J, Fuller J, Macaulay M, Hatz B, Jahoor A, Powell W, Waugh R (1997) Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor Appl Genet 95:714–722
Saghai-Maroof M, Soliman K, Jorgensen RA, Allard R (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci 81:8014–8018
Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR (2007) Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet 3:e133
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson LU (1994) Antioxidants and hormone-mediated health benefits of whole grains. Crit Rev Food Sci Nutr 34:473–497
Wang J-m, Yang J-m, Zhu J-h, Jia Q-j, Tao Y-z (2010) Assessment of genetic diversity by simple sequence repeat markers among forty elite varieties in the germplasm for malting barley breeding. J Zhejiang Univ Sci B 11:792–800
Zhang J, Liu X (2006) Descriptors and data standard for barley (Hordeum vulgare L.). Chinese Agricultural Press, Beijing
Zhang D, Zhang H, Wang M, Sun J, Qi Y, Wang F, Wei X, Han L, Wang X, Li Z (2009) Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor Appl Genet 119:1105–1117
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31101149), Natural Science Foundation of Zhejiang Province (Grant No. Y3110574), Public Benefit Technology Applied Research Project of Zhejiang Province (Project No. 2012C22029), China Agriculture Research System (CARS-05) and the Key Research Foundation of Science and Technology Department of Zhejiang Province of China (2012C12902-2).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hua, W., Zhang, X., Zhu, J. et al. A study of genetic diversity of colored barley (Hordeum vulgare L.) using SSR markers. Genet Resour Crop Evol 62, 395–406 (2015). https://doi.org/10.1007/s10722-014-0165-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-014-0165-7