Abstract
Recent advances in next generation sequencing technologies make genotyping by sequencing (GBS) more feasible for molecular characterization of plant germplasm with complex and unsequenced genomes. We used a GBS protocol consisting of Roche 454 pyrosequencing, genomic reduction and advanced bioinformatics tools to analyze genetic diversity of 24 diverse yellow mustard accessions. One and one half 454 pyrosequencing runs generated roughly 1.2 million sequence reads totaling about 392 million nucleotides. Application of the computational pipeline DIAL identified 512 contigs and 828 SNPs. The BLAST algorithm revealed alignments of 214 contigs with the sequences reported in NCBI nr/nt database. Sanger sequencing confirmed 95 % of 41 selected contigs and 94 % of 240 putative SNPs. The 454 scored SNPs were highly imbalanced among assayed samples. Diversity analysis of these SNPs revealed that 26.1 % of the total variation resided among landrace, cultivar and breeding lines and 24.7 % between yellow- and black-seeded germplasm. Cluster analysis showed that the black-seeded accessions were largely clustered together and the breeding lines were grouped with known origin. Computer simulation was performed to assess the impact of 454 SNPs missing and revealed considerable changes in allelic count, bias in detection of genetic structure, and large deviations from the expected genetic-distance matrix. These findings are useful for parental selection consideration in yellow mustard breeding, and our detailed analyses help illustrate the utility of GBS in genetic-diversity analysis of plant germplasm, particularly for genetic-relationship assessment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genotyping by sequencing (GBS) has recently emerged as a promising genomic approach for exploring plant genetic diversity on a genome-wide scale (Huang et al. 2009; Elshire et al. 2011; Fu and Peterson 2011; Poland and Rife 2012), thanks to the advances in next generation sequencing (NGS) technologies (Bräutigam and Gowik 2010; Metzker 2010). Generally, the GBS approach starts by reducing genome complexity with restriction enzymes, barcoding enzyme-cut genomic DNAs with indexed adaptors, multiplex-sequencing the barcoded DNA fragments in high-throughput NGS platforms, followed by a bioinformatics analysis of indexed sequence reads to identify genetic variants and a genetic diversity analysis of assayed samples. This approach requires no prior sequencing of plant genome, provides direct genotyping of plants with complex genomes without prior SNP discovery, and is capable of producing high-density, low-cost genotype information. These advantageous GBS features make it well suited as an informative molecular characterization tool, which can efficiently tap the large volume of ex situ plant germplasm conserved in world genebanks (FAO 2010), given that most conserved ex situ germplasm accessions have complex, unsequenced genomes (Fu and Peterson 2011).
Genotyping by sequencing has been shown to be a valid tool for genetic mapping (Baird et al. 2008; Elshire et al. 2011; Poland et al. 2012a), genomic selection for breeding (Poland et al. 2012b), and genetic-diversity studies (Fu and Peterson 2011, 2012; Lu et al. 2013). For example, Fu and Peterson (2011) applied the Roche 454 GS FLX Titanium technology with reduced genome representation and advanced bioinformatics tools to analyze the genetic diversity of 16 diverse barley landraces, discovered 2,578 contigs and 3,980 SNPs, and confirmed a key geographical division in the cultivated barley gene pool. Lu et al. (2013) developed a network-based SNP discovery protocol to enhance the diversity analysis of 540 switchgrass plants sampled from 66 populations and revealed informative patterns of genetic relationship with respect to ecotype, ploidy level and geographic distribution. These studies are encouraging for the use of GBS for genetic diversity analysis of plant germplasm. Still, very few studies have been conducted specifically to assess the effectiveness and informativeness of GBS for genomic characterization of plant germplasm (Fu and Peterson 2012).
Since the first effort to genotype at the genome-wide scale in rice (Huang et al. 2009), several GBS protocols have been developed for organisms without sequenced genomes and for improvement of genome coverage by reduced representation library (RRL), or genomic reduction, sequencing (Altshuler et al. 2000). RRLs have been widely applied to reduce the complexity of plant genomes (cf. Gore et al. 2009; Maughan et al. 2009; Deschamps et al. 2010; Hyten et al. 2010). They are constructed by means of a restriction enzyme (RE) digest followed by fragment-size selection, and allow for sampling diverse, but identical, genomic regions from several individuals (Altshuler et al. 2000). Widely applied GBS protocols are involved in the sequencing of either selected DNA fragments adjacent to RE cut sites (Baird et al. 2008; Elshire et al. 2011; Poland et al. 2012a; Peterson et al. 2012) or RE-cut DNA fragments of a certain length (Maughan et al. 2009; Fu and Peterson 2011). However, these GBS protocols usually display non-uniform genomic sampling (Beissinger et al. 2013), which may reduce overall coverage.
Another feature of the GBS approach is the generation of a large amount of missing SNP data per sample. Such missing data are expected as the selected DNA fragments may not be present and sequenced in each sample due to the loss or addition of a restriction site by mutation and/or biases in fragment selection and NGS sequencing (Nielsen et al. 2011), and because SNP-identification errors may occur due to variation in sequence-read depth, gene duplication and copy, and SNP-algorithm sensitivity (Pool et al. 2010). Efforts have been made to minimize missing data by sequencing to higher read depths (Poland et al. 2012a) or by filling the “blanks” with imputation (Marchini et al. 2007; Poland et al. 2012b). However, little is known about the impact of missing data on subsequent genetic analyses (Poland et al. 2012b), particularly on the analysis of genetic diversity (Fu and Peterson 2012).
Yellow mustard (Sinapis alba L.; 2n = 24; 1 pg = 0.50) has been cultivated as a condiment crop for millennia (Hemingway 1995; Bennett et al. 1982) and widely grown as a major specialty crop in the western Canadian prairies since the 1940s (Downey and Rakow 1995). Canadian breeding has been focusing on seed-yield increase since the 1950s (Downey and Rakow 1995; Katepa-Mupondwa et al. 2005). As yellow mustard is an obligate outcrossing species (Olsson 1960), recurrent selection has been a widely used breeding method (Cheng et al. 2012), but it is not always effective for seed yield due to low heritability. To enhance breeding efforts, diverse accessions of yellow mustard germplasm have been collected from different parts of the world, and 132 yellow mustard accessions are now maintained at Plant Gene Resources of Canada (PGRC) at Saskatoon. Clearly, exploring these yellow mustard accessions for genes that contribute to seed yield requires detailed characterization. Genetic-diversity analyses could generate useful information for understanding this species’ genetic variability and its patterns in germplasm originating from different countries, which could provide effective guidance on the selection of diverse parents for yield and quality improvements. A previous analysis of yellow mustard germplasm based on 134 AFLP markers revealed a large AFLP difference (15.6 %) residing between the yellow- and brown-seeded accessions, but only 6.2 % difference observed between the cultivar and landrace accessions (Fu et al. 2006). Clearly, a wider genome sampling with more informative genetic markers is warranted to assess genetic diversity for current yellow mustard breeding.
The objectives of our study were to (1) apply the GBS approach to identify contigs and SNPs from 24 diverse yellow mustard germplasm accessions by using 454 pyrosequencing via genomic reduction and the DIAL computational pipeline (Ratan et al. 2010), (2) use resulting 454 SNP data to analyze these accessions’ genetic diversity, and (3) employ computer simulation to assess the effects of missing SNPs on our genetic diversity analysis. It is our hope that this research effort will also help provide an assessment of the utility of GBS for genetic diversity analyses of plant germplasm.
Methods
Plant materials and DNA extraction
Twenty-four yellow mustard accessions, including four cultivars, 10 landraces and 10 inbred breeding lines (Table 1 and Table S1), were used in the study. The landraces and cultivars are open-pollinated populations originated from different countries. The 10 inbred breeding lines were developed at Agriculture and Agri-Food Canada Saskatoon Research Center. Y1352-9, Y1476-1, Y1485-5 Y1495-2, Y1355-2, Y1487 and Y1492 are inbred lines produced by inbreeding of different open-pollinated plants of the cultivar Andante (Table S1). Y1354-2 is the doubled haploid line SaMD3 produced by Bundrock (1998). Y1486-2 is derived from inbreeding of a Russian landrace. Y1354-7 was produced by seven generations of inbreeding of the F1 plant between the cultivar Sabre and the Svalöf high oil line (Todd Olson, personal communication with B. F. Cheng, 2010). About 10 seeds were randomly chosen from each selected accession. Plants were grown from seed for 3–4 weeks in a greenhouse at the Saskatoon Research Centre. Young leaf tissue from individual plants of each accession was collected, freeze-dried, and stored at −20 °C. DNA from one plant per accession was extracted from 15 mg of freeze-dried tissue with a DNEasy Plant Mini kit (Qiagen, Mississauga, ON, Canada) following the manufacturer’s instructions, quantified by using a Thermo Scientific Nanodrop 8000 (Fisher Scientific, Ottawa, ON, Canada), and adjusted to 100 ng/μl in Qiagen AE buffer (10 mM Tris–HCl, 0.5 mM EDTA, pH 9.0).
Genome reduction and barcoding
Genomic reduction and multiplex-identifier (MID) barcoding of the yellow mustard samples were conducted following the method of Maughan et al. (2009) by using the same sourced reagents and supplies where possible. EcoRI and BfaI adaptors and barcoded PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). The 24 samples were divided into two pools (Table 1). All samples were digested with EcoRI and BfaI. BfaI- and biotin-modified EcoRI-adaptors were ligated onto the digested fragments. The ligation reactions were cleaned on Chroma Spin +TE-400 columns (Clontech, Mountain View, CA, USA) following the manufacturer’s instructions. Fragments with the biotin-modified EcoRI-adaptor were selected by using streptavidin coated paramagnetic beads (Dynabeads M-280; Invitrogen, Burlington, ON, Canada) according to the manufacturer’s instructions.
Twenty-four unique Roche 454 RLMID barcodes were selected and used to identify 24 samples (Table 1). Paramagnetic beads with bound, digested DNA fragments were used as templates for PCR by using primers specific to the EcoRI- and BfaI-adaptors, containing a specific MID barcode for each sample in each pool. The PCR method was followed from Maughan et al. (2009) by using the Clontech HF2 chemistry and a C1000 thermocycler (BioRad, Mississauga, ON, Canada). Between four and six replicates of each PCR reaction were carried out, and a 3 μl sample from each was separated on a 1.5 % agarose gel to confirm amplification. Successful amplicons for each sample were bulked together and concentrated by evaporation in a vacuum centrifuge to approximately 35 μl. Individual samples were separated on a 1.5 % agarose gel for 5 h at 60 V. A gel fragment from each sample between 400 and 600 bp based on the New England Biolabs 2-Log ladder (Pickering, ON, Canada) was excised and cleaned by using the Qiaquick Gel Extraction kit (Qiagen, Mississauga, ON, Canada). Samples were eluted in 35 μl of one-third concentration Qiagen EB (3.33 mM Tris; pH 8.5) and quantified with the Thermo Scientific Nanodrop 8000. Individual samples were concentrated by evaporation in a vacuum centrifuge, re-quantified, and adjusted to 50 ng/μl with water and 1 mM EDTA pH 8.0, so that the final salt concentration did not exceed 10 mM Tris and 1 mM EDTA. Each pool was prepared with 200 ng of each of eight individual samples for a total of 1,600 ng at 50 ng/μl.
Pools were submitted to the DNA Technologies Laboratory at the Canadian National Research Council, Saskatoon, Saskatchewan, Canada, and sequenced on a full Roche 454 PicoTiterPlate (PTP) by using the Roche 454 GS FLX instrument with Titanium chemistry. An extra run was also made on a half PTP plate (i.e., a quarter PTP for each pool) by using Roche 454 GS FLX+ instrument with Titanium chemistry.
Generation of contigs and SNPs
DNA reads were combined from two 454 pyrosequencing runs and separated into sample-specific SFF files according to MID barcode based on the Roche Newbler SFF tools, followed by the removal of the forward and reverse adaptor sequences. Contig generation and SNP detection were performed with the DIAL pipeline (Ratan et al. 2010). The pipeline adds the SFF file of each sample and performs a completely automatic call of SNPs from all added SFF files in a Linux system. However, it requires both the input on the expected length of a target genome to identify contigs from all added SFF files and the version of Roche Newbler, as it is dependent on the Newbler’s gsAssembler to assemble the reads into the identified contigs for SNP identification. Thus, DIAL was trained for different versions of Newbler and variable lengths of target genome from 100 Mbp to 50 kbp. The final analysis was made by using Newbler v2.0.01.14 and an expected genome size of 300 kbp to generate the maximum numbers of contigs with SNPs for the 24 samples. All the training analyses generated an unrealistically low yield of 1–2 SNPs in the output file snps.txt due to the use of highly stringent filters for SNP calling. However, the pipeline also generated an output file report.txt collecting all the assembled contigs with the length and supporting reads, the position of the variant alleles, the number of reads supporting the allele, and the quality value of the reads at that position. Several specific Perl scripts were written to extract contigs and SNPs from report.txt into separate files for validation and for data report and analysis, and these custom-built Perl scripts are available upon request to the corresponding author.
Contig annotation
Searches by basic local alignment search tool (BLAST) (Altschul et al. 1990) for all identified contigs were made by using two approaches to provide some level of validation and gene ontology (GO) annotation on the contig sequences. The first was to conduct BLAST searches directly against the NCBI nr/nt protein database in the NCBI website (http://www.ncbi.nlm.nih.gov/). The second was to employ the program Blast2GO (Conesa et al. 2005) against the NCBI nr protein database. Specifically, the applied annotation parameters were a pre-e-value-Hit-Filter (10−6), annotation cut-off threshold (55) and GO weight (5). Blast2GO uses BLAST to find similar sequences (potential homologs) for one or several input sequences, extracts all GO terms associated to each of the obtained hits, and returns an evaluated GO annotation for the query sequence(s).
Contig and SNP validation
A random set of 41 contigs was selected for validation with Sanger sequencing (SS) based on three randomly selected samples (SA44, SA115, Y1476-1). The contig selection considered only the variable SNP count and contig length, not the BLAST search results. The PCR primers for 41 contigs were designed by using Primer3 (v.0.4.0) (Rozen and Skaletsky 2000). The conditions for PCR were: 1× KAPA 2G Buffer A containing 1.5 mM MgCl2 (KAPA Biosystems, Woburn, MA, USA), 1× KAPA Enhancer 1, 0.2 mM each dNTP, 0.4 pmol/μl each forward and reverse primers, 100 ng of the same genomic DNA template samples as used above for NGS, and 0.5 U KAPA 2G Robust polymerase in a final volume of 25 μl; touchdown PCR cycled at 95 °C for 3 min followed by 10 cycles of 95 °C for 10 s, 60 °C decreasing 0.5 °C per cycle for 15 s, 72 °C for 30 s followed by 25 cycles of 95 °C for 10 s, 55 °C for 15 s, 72 °C for 20 s, followed by a final extension of 72 °C for 30 s. A 3 μl sample of each PCR product was separated on 1.5 % agarose for 2 h at 120 V. Two primer sets amplified no or multiple products. For the remaining 39 primer sets, their PCR products were cleaned following the method outlined by Rosenthal et al. (1993) and submitted to the DNA Technologies Laboratory at the Canadian National Research Council, Saskatoon, for Sanger sequencing.
Forward and reverse Sanger sequences from each sample were assembled and aligned with Sequencher v.5.0 (GeneCodes, Ann Arbor, MI, USA), then aligned against the consensus sequence generated from 454 pyrosequencing for each contig by using Muscle v.3.6 (Edgar 2004), and proofread by hand. The putative 454 SNPs were checked with the Sanger sequences, where sample data were available, and additional SNPs and indels from the SS, if any, were also identified.
Comparative SNP identification by Roche Newbler
Roche Newbler GS Reference Mapper software (version 2.6p1 supplied by Roche in November, 2011) was also run for all Roche 454 sequence reads generated for this study against the 39 contigs confirmed by SS. The software called all sequence differences between the sequences of 39 contigs and assayed samples, including SNP and indel, and stored them in the file 454AllDiffs.txt. A specific Perl script was written to extract genetic variants from 454AllDiffs.txt and to compare them to those identified by the SS and DIAL pipeline.
Diversity analysis
The 454 SNP data obtained from the DIAL pipeline were analyzed for each sample by counting the total putative SNPs, the heterozygous SNPs, and the SNPs that were undetected in the sample due to insufficient sequence reads. As the 454 SNP data are highly imbalanced, a random permutation rest was made on the pairwise sample differences in SNP count. This was done by a random permutation of the 454 SNPs (including missing ones) per locus over the 24 samples and repeat of the permutation for all the loci, followed by the SNP count for each sample from the permuted 454 SNP data and the calculation of the permuted pairwise sample differences in SNP count. This process was run 10,000 times to calculate the proportion of runs in which the permuted pairwise sample difference was larger or smaller than (depending on the sign of) the observed pairwise sample difference in SNP count, giving the significant level of the test for each pairwise sample difference in SNP count. The random permutation was performed with a custom R script within R version 2.15 (R Development Core Team 2011) that is available upon request.
An analysis of molecular variance (AMOVA) was performed with Arlequin version 3.01 (Excoffier et al. 2005) on the 454 SNP data to quantify the genetic variation present among various groups of samples (landrace, cultivar, and breeding line; yellow- and black-seeded groups). To assess the impact of missing SNPs on the variation partition, the original SNP data were re-coded with 1 for a missing SNP and 0 for an available SNP for each locus and sample (ignoring the nucleotide information), and AMOVA was performed on the re-coded data based on the above group structures.
The genetic relationships of the 24 yellow-seeded mustard samples were determined with three different approaches for comparison. The first was to generate a neighbor-joining dendrogram by using NTSYSpc 2.01 (Rohlf 1997) based on the dissimilarity matrix of the available putative SNPs. The second was to generate a neighbor-joining tree with PAUP* (Swofford 1998) and display it by using MEGA5 (Tamura et al. 2011). The third was to generate a distance-based NeighborNet (Bryant and Moulton 2004) of the 24 samples by using the SplitsTree4 (Huson and Bryant 2006). To assess the impact of missing SNPs on sample clustering, the re-coded data for missing versus existing SNPs were used to determine the sample genetic relationships by following three approaches mentioned above.
Computer simulation
To understand the effects of missing SNPs on the genetic diversity analysis, a Monte Carlo computer simulation was performed based on available 454 SNP data with an average of 73 % SNPs missing per sample. Ten scenarios of missing SNPs were considered: completely random missing SNPs at the missing levels of 5, 20, 35, 50, 65, 80, and 95 %, completely random with equal missing SNP level of 73 % for each sample (73e), randomly matched individually with the existing missing SNP level of 73 % (73r), and randomly matched individually with the existing missing level and pattern of 73 % (73f). Each simulation started with a generation of a full SNP data set by randomly allocating four nucleotides (A, C, G, T) based on the observed nucleotide frequencies at each locus (available from the 454 SNP dataset) to the 24 samples and repeating for 828 loci. Then, a data set with missing SNPs was generated for each missing scenario by selecting randomly from, or (for the 73f scenario) matching observed patterns of missing data with, the simulated full SNP data set. Next, a diversity analysis was performed on both full and missing SNP data sets to estimate four diversity parameters (as described in the following paragraph). This process was repeated 5,000 times, and the mean and standard deviation of the parameter estimates were obtained.
Our simulation examined four diversity parameters: allelic counts for two groups of alleles (a tail group of alleles of frequencies smaller than 0.1 and a middle group of alleles of frequencies from 0.45 and 0.55), the probability of detecting a population genetic structure under missing SNPs, and the congruency between two distance matrices representing 24 samples with and without missing SNPs. The AMOVA algorithms (Excoffier et al. 1992) were used to estimate the sum of squared differences among (SSA) and within (SSW) three groups of samples (also see AMOVA analysis above), and the number of the simulation runs where SSA is larger than SSW provided the estimate of the probability of detecting a population genetic structure. Pairwise sample SNP dissimilarities were calculated following Fu (2006) from full or missing SNP data, and two dissimilarity matrices were used to estimate the normalized Mantel correlation coefficient (Mantel 1967). The simulations were conducted with a custom R script within R version 2.15 (R Development Core Team 2011) that is available upon request.
Results
The workflow for GBS of yellow mustard germplasm was summarized in supplemental Fig. S1. The GBS protocol was relatively straightforward and cost-effective for generating a large amount of genotype data. Specifically, the GBS application generated 1.2 million passed reads with about 392 Mbp of DNA sequence from 1.5 full 454 runs of the 24 samples (Table 2). The samples for the pool A had considerably fewer reads than did those in the pool B over the runs. The sample Y1476-1 had the most passed reads (392,797) among the assayed samples. In contrast, the sample SA88 had the fewest passed reads (11,649) (Table 1).
Training DIAL with the original Newbler v2.0.01.14 revealed that the numbers of identified contigs and SNPs increased with an applied genome size decreasing from 100 Mbp and reached a maximum number of 512 contigs and 828 tentative SNPs with a genome size of 300 kbp or smaller. For example, with 5 Mbp, 490 contigs and 800 SNPs were identified, and with 1 Mbp, 501 contigs and 820 SNPs were found. Training with the newer version (2.6p1) revealed that the maximum numbers of both contigs and SNPs obtained become much smaller. For example, the training with the newer version identified only 312 contigs and 558 SNPs at the genome size of 300 kbp.
A total of 512 contigs were detected from 6,008 (0.5 %) passed reads (Table 2; supplemental Table S2). Effectively, only 6.9 % of the sequence bases were used to identify the contigs and 0.05 % bases contributed to the 512 contigs found. Note that contigs that were shorter than 100 bp or had fewer than six sequence reads were not reported. The number of reads per contig ranged from 2 to 69 and averaged 12. Only 21 contigs had two to four reads. The contig length ranged from 100 to 883 bp with an average of 382.3 bp and a median of 399.5 bp. There were 192 contigs without any SNPs and 320 contigs with up to 12 SNPs. We identified 828 putative SNPs on the 320 contigs from the 24 samples. For those 320 contigs, the average and median numbers of putative SNPs per contig were 2.6 and 2, respectively.
The BLAST search of 512 contigs showed that 214 (41.8 %) contigs matched sequences reported in the NCBI nr/nt database (results not shown). The BLAST search through Blast2GO software revealed that 199 (38.9 %) contigs were associated with reported gene annotations (Table S2). Across these hits the matches were associated with at least 27 plant species, and the four species with more than 10 hits were Arabidopsis thaliana (167), Arabidopsis lyrata (59), Brassica oleracea (14) and Brassica rapa (11) (Fig. S2).
The 41 primer sets designed on 41 (8 %) contigs that were selected for validation via SS confirmed 39 (95.1 %) contigs (Table 2), while two primer sets did not yield sequence. Note that neither of these two contigs had no hits in both BLAST searches (Table S2). SS of the three samples taken from the 39 confirmed contigs validated 226 (87.3 %) of the 259 putative SNPs identified by the DIAL pipeline. If 19 SNPs that resided outside of the flanking primers on the 16 contigs were excluded, the SNP validation rate was 94.2 % (=226/240). The 14 non-validated SNPs were either due to the lack of Sanger sequence data (6) or monomorphism among three samples (8). Also, the SS revealed 240 (93 %=240/259) more SNPs and indels than the DIAL prediction did on the same contigs (Table 2). Specifically, the SS detected 207 more SNPs on 30 contigs and 33 new indels on 15 contigs (Table S3).
The effective rate of SNP discovery from both Sanger and 454 pyrosequencing in the confirmed 39 contigs ranged from 6.6 to 11.9 SNPs and indels per contig with read lengths of 253–569 bp, or 3.1 SNPs and indels per 100 bp (=466 SNPs and indels/14,990 bp for the 39 contigs) (Table 2). More specifically, the effective SNP discovery rate was 2.89 SNPs/100 bp (=433 SNPs/14,990 bp), and the effective indel discovery rate was 0.22 indels/100 bp (=33 indels/14,990 bp) (Table S4).
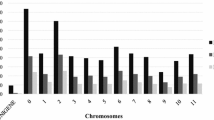
The Roche 454 GS Reference Mapper software revealed 1,028 genetic variants among the 24 samples on the 39 confirmed contigs, of which 458 were classified as “High-Confidence” by the software (Fig. 1). Among the 1,028 genetic variants, 56 were the same as those of the 259 variants identified by the DIAL pipeline and 162 (=56 + 106) as those of the 499 variants identified by the SS (Fig. 1A). Considering only those 458 “High-Confidence” genetic variants, 37 were the same as those of the 259 variants identified by the DIAL pipeline and 106 (=37 + 69) as those of the 499 variants identified by the SS (Fig. 1B). In contrast, 226 (=56 + 170) of those 259 genetic variants identified by the DIAL pipeline matched those of the 499 variants identified by SS (Fig. 1A).
Shared genetic variants identified by the DIAL pipeline and GS Reference Mapper software in comparison with the Sanger sequence variants on 39 confirmed contigs from three yellow mustard samples. A All the variants detected by GS mapper and B all the variants with “High-Confidence” detected by GS mapper
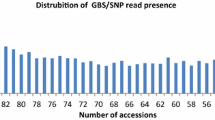
The number of contigs and SNPs identified for each sample varied substantially, ranging from 13 to 233 contigs with an average of 88 and from 29 to 524 SNPs with an average of 225 (Table 1). Similarly, the percentages of the observed heterozygotes and missing SNPs also varied greatly, ranging from 0 to 24 % with an average of 4.6 % for heterozygotes and from 36.7 to 96.5 % with an average of 73 % for missing SNPs. Such variation was largely associated with the number of passed reads per sample, as revealed by simple linear regression analyses (r 2 = 0.43, p < 0.0001 for contigs, r 2 = 0.31, p < 0.004 for SNP, and r 2 = 0.31, p < 0.004 for missing SNPs). The observed heterozygotes were not associated with the number of passed reads, but highly related to the number of contigs and SNPs identified (r 2 = 0.49, p < 0.0001 for contigs and r 2 = 0.57, p < 0.0001 for SNPs). However, the average percentage of observed heterozygotes for breeding lines was lower (2.3 %) than those for cultivars (7.3 %) or landraces (5.8 %), and the highest percentage of observed heterozygotes was observed in the landrace sample SA115 (24 %). Most of the pairwise sample differences in SNP count were statistically significant, as revealed by the random permutation tests (Table 3). For example, the SNP count difference (32) between samples SA12 and SA23 was significant at p < 0.05, but the SNP count difference (10) between samples Y1494-2 and Y1495-2 was not significant.
The AMOVA that considered the missing data revealed 26.1 % SNP variation present among landraces, cultivars and breeding lines and 24.7 % SNP variation residing between samples with black and yellow seeds (Table 4). Also, based on the group-specific proportion of SNP variation (gFst), it appeared that the yellow-seeded mustards had more SNP variation (gFst = 0.24) than did the black-seeded mustards (gFst = 0.26). Similarly, the cultivar samples seemed to display the most SNP variation (gFst = 0.24), followed by the landrace (gFst = 0.26) and breeding line (gFst = 0.27) samples, although these gFst differences may not be statistically significant. Furthermore, variance analysis with respect to missing versus existing SNP data revealed negative group-wise variances due to sampling correction in the calculation (Table 4), suggesting that missing SNPs had some negative effects on SNP variance estimation.
The genetic relationships of the 24 yellow mustard samples were illustrated by dendrogram, NJ tree, and NeighborNet in Fig. 2. It is clear that the black-seeded mustard samples were genetically distinct, with five of the six samples locating in the bottom of the dendrogram, while one was separated apart in the yellow-seeded mustard groups (Fig. 2A). The 10 breeding lines clustered in five separate groups, and two breeding lines were associated with black-seeded germplasm (Fig. 2A). These relationships are essentially the same as those observed in the NJ tree and NeighborNet, although the latter had more resolution. For example, the NJ tree showed more clearly the genetic distinctness of the two samples, SA88 and SA100 (Fig. 2B), while the NeighborNet displayed more articulations among the breeding lines than those among the black-seeded samples (Fig. 2C). Also, seven of the 10 breeding lines originated from the cultivar Andante were clustered together in the NJ trees (Fig. 2B), which slightly differs from those displayed in the dendrogram (Fig. 2A). Mantel tests on associations between two matrices of the pairwise sample genetic distances generated by the three clustering methods and of the pairwise sample differences in the proportional missing SNPs were not statistically significant (results not shown). However, further analysis with respect to missing versus existing SNPs revealed that increased missing SNPs resulted in smaller genetic differences of a sample from the other samples, as shown in Fig. S3. For example, the sample SA115, with 37 % missing SNPs, had much longer branch than did sample SA23, having 94 % missing SNPs.
Genetic relationships of the 24 yellow mustard samples as revealed by cluster analyses of 828 identified SNPs. A The neighbor-joining (NJ) dendrogram obtained by using NTSYSpc software. B The NJ tree generated from PAUP*software. C The NeighborNet generated from SplitsTree4 software. Seven accessions with black seeds are additionally labeled with a star and highlighted with filled circle nodes in the NJ tree. Seven breeding lines originated from the Andante cultivar are highlighted in the NJ tree
Our computer simulation revealed three sets of interesting results. First, the group of alleles with frequencies from 0.45 to 0.55 increased up to 40 % with increased levels of missing SNPs (Fig. 3A). Variation among samples of missing SNPs increased further for the mentioned group of alleles, as reflected in three specific scenarios (i.e., 73e, r, and f). Interestingly, more variation was observed for the group of alleles with frequencies <0.1 (Fig. 3B). When the missing level was <50 %, the count for this group of alleles was smaller than the true count, but when the missing level was >50 %, the count for this group of alleles was much larger than expected. More variation was also observed with respect to variation in patterns of missing SNPs (i.e., for 73e, r, and f).
Results of computer simulation. Ten scenarios of missing SNPs: 5, 20, 35, 50, 65, 80, 95, 73e, 73r, 73f, are described in the text. A Ratio of simulated versus true counts for group alleles of frequencies from 0.45 to 0.55 (i.e., Countgs and Countgt, respectively). B Ratio of simulated versus true counts for group alleles of frequencies smaller than 0.1. C The probability that a genetic structure was detected (SSA > SSW) when there was no genetic structure. D Normalized Mantel correlation coefficients between one distance matrix calculated from a full SNP data and a distance matrix from data with various levels of missing SNPs
Second, the probability of detecting genetic structure among the 24 samples, when in reality there was no genetic structure, deviated little from our expectations (0.5) for eight scenarios representing random distributions of missing SNPs, but was close to 1 when the missing data were not completely random (i.e., for 73r and 73f) (Fig. 3C).
Third, the normalized Mantel correlation coefficients for pairwise distances among the 24 samples with and without missing SNPs decreased with increasing proportions of missing SNPs. More missing SNPs helped create genetic-distance matrices that varied more widely from the expected genetic-distance matrix. Variation in patterns of missing SNPs among different samples would further enlarge deviations from the expected genetic-distance matrix.
Discussion
This study represented the first attempt to characterize yellow mustard germplasm based on a GBS protocol. Our GBS application not only generated a novel set of genomic resources useful for further genomic analysis of yellow mustard, but also provided a large genotypic data set for genetic-diversity analyses of the assayed samples. The diversity analysis of the 454 SNP data revealed 26.1 % SNP variation resided among landrace, cultivar and breeding lines and 24.7 % between yellow- and black-seeded types. The cluster analysis showed that black-seeded germplasm largely grouped together; breeding lines were grouped with known origin; and two of them were associated with black-seeded germplasm.
Given an estimated genome size of 500 Mbp for yellow mustard (Bennett et al. 1982), this GBS application generated a smaller set of genomic resources (i.e., 512 contigs and 828 SNPs) than did our previous GBS efforts in barley (2,578 contigs and 3,980 SNPs; Fu and Peterson 2011) and flax (713 contigs and 1,067 SNPs; Fu and Peterson 2012). Such low genomic output was unexpected for an outcrossing species, but may reflect that the combination of EcoRI and BfaI REs may have not yielded sufficient restriction fragments of an appropriate size in this genome and that other combinations of 6- and 4 bp-recognizing enzymes may be more effective (Peterson et al. 2012). Interestingly, the average level of the missing data per sample (72.9 %) was compatible with those reported for barley (70.1 %; Fu and Peterson 2011) and flax (68.9 %; Fu and Peterson 2012). Given these three studies employed the same RE, this comparison suggests that the magnitude of missing SNPs may be highly associated with specific RE combinations used.
The rates of validation on both 454 contigs and SNPs by SS were high (95 % and 94, respectively) and compatible with those previously reported in barley (Fu and Peterson 2011) and flax (Fu and Peterson 2012). However, the effective rate of SNP and indel discovery in yellow mustard was higher than those observed in barley and flax; this is expected given that yellow mustard is an outcrossing crop. Interestingly, the DIAL computational pipeline yielded more accurate SNP identification than did the Roche 454 GS Reference Mapper software (Fig. 1). This finding, along with those previously reported (Fu and Peterson 2012), suggests caution in selection of a computational pipeline to identify SNPs.
The diversity analysis of the 454 SNP data revealed higher levels of SNP variation residing among landrace, cultivar and breeding lines (26.1 %) and between yellow- and black-seeded groups (24.7 %) than those reported on 127 accessions based on AFLP markers (20.9 and 15.6 %, respectively; Fu et al. 2006). Given the outcrossing nature of yellow mustard and the homogeneity of the breeding lines used, the levels of SNP variation should be smaller. This discrepancy may reflect the impact of missing data, as evidenced by the negative variance estimates obtained from sampling correction (Table 3; Bird et al. 2011), but the exact level of impact remains unknown. Also, the cultivars appeared to display more SNP variation than the landraces. This unexpected result may be due to the biases of sampling outcrossing individuals for an accession and/or missing 454 SNPs.
The genetic relationships of the 24 yellow mustard accessions inferred by three clustering approaches (Fig. 2) were largely consistent with those patterns detected by AFLP markers (Fig. 1 of Fu et al. 2006), where black-seeded accessions were genetically more distinct and largely clustered together. As shown in Fig. S3, missing data reduce the SNP differences of a sample against others, but such reduction did not seem to disturb the overall pattern of genetic relationships among assayed samples. To some extent, this is understandable, as an average 73 % missing SNPs per sample in this 454 SNP dataset means each sample still has an average of 225 SNPs available to estimate pairwise genetic distances with other samples. The resulting genetic distances based on so many SNPs should still be generally informative. More interestingly, the revealed genetic relationships help confirm the relatedness of the 10 breeding lines with known origin (Fig. 2B) and should be valuable for parental selection consideration in yellow mustard breeding. For example, the two breeding lines Y1476-1 and Y1352-9 of known origin were confirmed to be closely related, even with 37.7 and 85.9 % missing SNPs, respectively.
Without the complete 454 SNP data, it is impossible to perform an empirical assessment on the impacts of the resulting missing SNP data on current genetic-diversity analysis. However, our computer simulation helped shed some light on the effects of missing data of such magnitude on three major components of the diversity analysis (allelic richness, genetic structure, and genetic distance). Not surprisingly, missing data introduce a large bias in the estimation of allelic richness (Fig. 3A, B). The detection of genetic structure will be largely affected only if the missing data are not completely random (Fig. 3C). The magnitude and pattern of missing data can alter a genetic-distance matrix away from expectations. It is worth noting, however, that our simulation was preliminary and not exhaustive, using only the resulting 454 SNP frequencies and considering only three diversity components under a few scenarios of missing data. Further simulations are needed to comprehend the overall impacts on various diversity parameters estimations over broader scenarios of missing data including DNA fragment loss.
The present study did not apply any imputation procedures (Poland et al. 2012b) to improve genetic-diversity analysis, as imputation may not help much to reduce sampling bias in allelic richness estimation, particularly for species such as yellow mustard without a sequenced genome (Pool et al. 2010). However, imputation without a reference genome could theoretically reduce the sampling bias in genetic-distance estimation to enhance the inference of genetic relationships (YB Fu, unpublished results), and consequently explorations on the use of imputation without a reference genome may be worth pursuing. Also, our GBS protocol has not yet been optimized with respect to sample size, RE, or sequencing platform (Elshire et al. 2011), as our application was limited to a small sample of 24 accessions and considered only one pair of REs in a single sequencing platform. Thus, our assessment on GBS performance in genetic diversity analysis was not comprehensive.
In spite of these limitations, our findings are significant for genetic diversity analysis of plant germplasm, particularly considering 7.4 millions of underexplored ex situ germplasm accessions conserved worldwide (FAO 2010). Our first attempt generated a novel set of resources for future genomic analyses of yellow mustard. These detailed analyses help illustrate the utility of GBS in the genomic characterization of plant germplasm. Our GBS protocol is relatively straightforward, rapid and cost-effective, does not require a reference sequence, and can provide high-density genotype data for genetic-diversity analysis. Our diversity analysis of yellow mustard accessions revealed genetic relationships among elite breeding lines that may be valuable for parental selection for yellow mustard improvement.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407:513–516
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376
Beissinger TM, Hirsch CN, Sekhon RS, Foerster JM, Johnson JM, Muttoni G, Vaillancourt B, Buell CR, Kaeppler SM, de Leon N (2013) Marker density and read-depth for genotyping populations using genotyping-by-sequencing. Genetics 193:1073–1081
Bennett MD, Smith JB, Heslop-Harrison JS (1982) Nuclear DNA amounts in angiosperms. Proc R Soc Lond (Biol) 216:179–199
Bird CE, Karl SA, Smouse PE, Toonen RJ (2011) Detecting and measuring genetic differentiation. In: Koenemann S, Held C, Schubart C (eds) Phylogeography and population genetics in Crustacea, vol 19., Crustacean Issues SeriesCRC Press, Boca Raton, FL, pp 31–55
Bräutigam A, Gowik U (2010) What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol 12:831–841
Bryant D, Moulton V (2004) NeighborNet: an agglomerative algorithm for the construction of planar phylogenetic networks. Mol Biol Evol 21:255–265
Bundrock T (1998) Doubled haploidy in yellow mustard (Sinapis alba L.). MSc thesis. University of Saskatchewan, Saskatchewan, Saskatoon, Canada
Cheng B, Williams DJ, Zhang Y (2012) Genetic variation in morphology, seed quality and self-(in)compatibility among the inbred lines developed from a population variety in outcrossing yellow mustard (Sinapis alba). Plants 1:16–26
Conesa A, Gotz S, Garcia-Gomez JM et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Deschamps S, Rota ML, Ratashak JP, Biddle P, Thureen D, Farmer A, Luck S, Beatty M, Nagasawa N, Michael L et al (2010) Rapid genome-wide single nucleotide polymorphism discovery in soybean and rice via deep resequencing of reduced representation libraries with the Illumina genome analyzer. Plant Genome 3:53–68
Downey RK, Rakow G (1995) Mustard. In: Slinkard AE, Knott DR (eds) Harvest of gold: the history of field crop breeding in Canada. University of Saskatchewan, Saskatoon, pp 213–219
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
FAO (2010) The second report on the state of the world’s plant genetic resources for food and agriculture. FAO, Rome
Fu YB (2006) Redundancy and distinctness in flax germplasm as revealed by RAPD dissimilarity. Plant Genet Resour 4:117–124
Fu YB, Peterson GW (2011) Genetic diversity analysis with 454 pyrosequencing and genomic reduction confirmed the eastern and western division in the cultivated barley gene pool. Plant Genome 4:226–237
Fu YB, Peterson GW (2012) Developing genomic resources in two Linum species via 454 pyrosequencing and genomic reduction. Mol Ecol Resour 12:492–500
Fu YB, Gugel R, Katepa-Mupondwa F (2006) Genetic diversity of Sinapis alba germplasm as revealed by AFLP markers. Plant Genetic Resour 4:87–95
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL et al (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Hemingway JS (1995) The mustard species: condiment and food ingredient use and potential as oilseed crops. In: Kimber D, McGregor DI (eds) Brassica oilseeds production and utilization. CAB International, Wallingford, pp 373–383
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B (2009) High throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hyten DL, Song Q, Fickus EW, Quigley CV, Lim JS, Choi IY, Hwang EY, Pastor-Corrales M, Cregan PB (2010) High-throughput SNP discovery and assay development in common bean. BMC Genomics 11:475
Katepa-Mupondwa F, Raney JP, Rakow G (2005) Recurrent selection for increased protein content in yellow mustard (Sinapis alba L.). Plant Breed 124:382–387
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH et al (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9:e1003215
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39:906–913
Maughan PJ, Yourstone SM, Jellen EN, Udall JA (2009) SNP discovery via genomic reduction, barcoding, and 454-pyrosequencing in amaranth. Plant Genome 2:260–270
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31–46
Nielsen R, Paul JS, Albrechtsen A, Song YS (2011) Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12:443–451
Olsson G (1960) Self-incompatibility and outcrossing in rape and white mustard. Hereditas 46:241–252
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7:e37135
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5:92–102
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012a) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253
Poland J, Endelman J, Dawson J, Rutkoski J, Wu S, Manes Y, Dreisigacker S, Crossa J, Sanchez-Villeda H, Sorrells M, Jannink J-L (2012b) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 5:103–113
Pool JE, Hellmann I, Jensen JD, Nielsen R (2010) Population genetic inference from genomic sequence variation. Genome Res 20:291–300
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, http://www.r-project.org/
Ratan A, Zhang Y, Hayes VM, Schuster SC, Miller W (2010) Calling SNPs without a reference sequence. BMC Bioinform 11:130
Rohlf FJ (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software, Setauket, NY
Rosenthal A, Coutelle O, Craxton M (1993) Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res 21:173–174
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ, pp 365–386
Swofford DL (1998) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
The authors would like to thank Carolee Horbach for technical assistance in 454 pyrosequencing and data processing; Richard Gugel for assistance in germplasm sampling; Matthew Links for assistance with access to a Linux server; and two journal reviewers for constructive comments on the early version of the paper. This research was partly funded by Agriculture and Agri-Food Canada A-base Program to Y. B. Fu and by Growing Forward II, Agri-Innovation Program, Canada, Mustard 21 Canada, Inc., and the Agriculture Development Fund (ADF) of Saskatchewan to B. F. Cheng.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiment complies with the current laws of Canada in which it was performed.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Fu, YB., Cheng, B. & Peterson, G.W. Genetic diversity analysis of yellow mustard (Sinapis alba L.) germplasm based on genotyping by sequencing. Genet Resour Crop Evol 61, 579–594 (2014). https://doi.org/10.1007/s10722-013-0058-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-013-0058-1